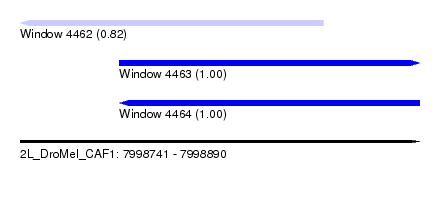
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,998,741 – 7,998,890 |
| Length | 149 |
| Max. P | 0.997641 |

| Location | 7,998,741 – 7,998,854 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.21 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -13.15 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7998741 113 - 22407834 ACUAAAUAUAGCCAUACAAUACGCACAGCGGAGACGCACAC-CAGGGUGCAUGACUGUGCUAGGCAUUGAGCUGUA---ACUCU-AGCUACAUG--CUAUCUGGCAACGCUGGCUUUUAA .........(((((........((((((((....)((((..-....))))..).))))))..(((((..(((((..---....)-))))..)))--))....(....)..)))))..... ( -34.50) >DroSec_CAF1 11041 99 - 1 ACUAAAUAUAGCCAUAAAACAGGCACAGCGGAGACGCACAU-CAGGGUAUAUGACUCUGCU--------------A---ACUAU-AGCUACAUG--UAAUCUGGCAACGCUGGGUUUUAA ..............((((((.(((...(((....)))...(-(((..((((((.....(((--------------(---....)-)))..))))--))..))))....)))..)))))). ( -27.50) >DroSim_CAF1 9391 96 - 1 ACUAAAUAUGGCCAUAA----CGCAUAGCGGAGACACACACCCAGGGUGUGUGACUCUGCU--------------A---ACUAU-AGCUGCAUG--CAAUCUGGCAACGCUGGCUUUCAA .........(((((...----.((.((((((((.((((((((...)))))))).)))))))--------------)---.....-.((.....)--).....(....))))))))..... ( -34.80) >DroEre_CAF1 9785 116 - 1 ACUAGUUAUGGCCUGACCAUAUGCUCAGCGGAGACGCACAC-CACGGAGCAGUACUUUGCUAGGCCUUUCGCGGAGCAGGGUGCAAG-CCCACG--CGAUCUGGCAGCGCUGGCUUUCAA ......(((((.....)))))(((((.(((....))).(..-...))))))((((((((((..((.....))..))))))))))(((-((..((--(.........)))..))))).... ( -42.90) >DroYak_CAF1 9618 98 - 1 ACUAGUUAUAGCCCCACAAUAUGCGCAGCGGAGACGCACAC-CGCGGUGCAUUACCUUGCUACAC--------------------AG-UCCACGCUCGAUCUGGCAACGCUGACAUUUAA ........((((........((((((.((((.(.....).)-))).))))))......))))..(--------------------((-(....(((......)))...))))........ ( -22.54) >consensus ACUAAAUAUAGCCAUACAAUACGCACAGCGGAGACGCACAC_CAGGGUGCAUGACUCUGCUA__C__________A___ACUAU_AGCUACAUG__CAAUCUGGCAACGCUGGCUUUUAA ........((((..........((((.(((....))).(......))))).......(((((.......................................)))))..))))........ (-13.15 = -12.87 + -0.28)


| Location | 7,998,778 – 7,998,890 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.07 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -14.66 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7998778 112 + 22407834 U---UACAGCUCAAUGCCUAGCACAGUCAUGCACCCUG-GUGUGCGUCUCCGCUGUGCGUAUUGUAUGGCUAUAUUUAGUACAUUUUCAGUGCCACCACAACAUAUGGCAC----AAUAA .---..((...((((((...(((((((.((((((....-..))))))....)))))))))))))..))((((....)))).........((((((..........))))))----..... ( -33.30) >DroSec_CAF1 11078 98 + 1 U---U--------------AGCAGAGUCAUAUACCCUG-AUGUGCGUCUCCGCUGUGCCUGUUUUAUGGCUAUAUUUAGUACAUUUUGAGUGCCACCCCAACAUAUGGCAC----AAUAA .---.--------------(((.(((.(((((......-)))))...))).)))(((((((((...((((...((((((......))))))))))....))))...)))))----..... ( -24.60) >DroSim_CAF1 9428 94 + 1 U---U--------------AGCAGAGUCACACACCCUGGGUGUGUGUCUCCGCUAUGCG----UUAUGGCCAUAUUUAGUACAUUUUCAGUGCCACC-CAACAUAUGGCAC----AAUAA .---(--------------(((.(((.((((((((...)))))))).))).))))....----...(((((((((...((((.......))))....-....))))))).)----).... ( -32.40) >DroEre_CAF1 9822 118 + 1 CCUGCUCCGCGAAAGGCCUAGCAAAGUACUGCUCCGUG-GUGUGCGUCUCCGCUGAGCAUAUGGUCAGGCCAUAACUAGUACAUUUCCAGUGCCUC-GCAACAUAUGGCAAAUAAAAUAA ..(((...((((..(((((..(...(((.(((((.(((-(.(.....).)))).)))))))).)..))))).......((((.......)))).))-))..(....)))).......... ( -35.60) >DroYak_CAF1 9652 99 + 1 ---------------GUGUAGCAAGGUAAUGCACCGCG-GUGUGCGUCUCCGCUGCGCAUAUUGUGGGGCUAUAACUAGUACAUUUUCAGUGCCUC-GCAACAUAUGGCAU----AAUAA ---------------((((.((.(((((.((..(((((-((((((((.......))))))))))))).((((....)))).......)).))))).-)).)))).......----..... ( -32.70) >consensus U___U__________G__UAGCAAAGUCAUGCACCCUG_GUGUGCGUCUCCGCUGUGCAUAUUGUAUGGCUAUAUUUAGUACAUUUUCAGUGCCACCGCAACAUAUGGCAC____AAUAA ....................(((.(((.((((((.......))))))....))).)))..........(((((((...((((.......)))).........)))))))........... (-14.66 = -15.10 + 0.44)

| Location | 7,998,778 – 7,998,890 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.07 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7998778 112 - 22407834 UUAUU----GUGCCAUAUGUUGUGGUGGCACUGAAAAUGUACUAAAUAUAGCCAUACAAUACGCACAGCGGAGACGCACAC-CAGGGUGCAUGACUGUGCUAGGCAUUGAGCUGUA---A .....----(((((((........)))))))...............((((((((.....((.((((((((....)((((..-....))))..).)))))))).....)).))))))---. ( -35.70) >DroSec_CAF1 11078 98 - 1 UUAUU----GUGCCAUAUGUUGGGGUGGCACUCAAAAUGUACUAAAUAUAGCCAUAAAACAGGCACAGCGGAGACGCACAU-CAGGGUAUAUGACUCUGCU--------------A---A ....(----(((((...((((...(((((.....................)))))..))))))))))(((....)))....-((((((.....))))))..--------------.---. ( -31.00) >DroSim_CAF1 9428 94 - 1 UUAUU----GUGCCAUAUGUUG-GGUGGCACUGAAAAUGUACUAAAUAUGGCCAUAA----CGCAUAGCGGAGACACACACCCAGGGUGUGUGACUCUGCU--------------A---A ....(----(.(((((((.(((-(...(((.(....)))).)))))))))))))...----....((((((((.((((((((...)))))))).)))))))--------------)---. ( -38.40) >DroEre_CAF1 9822 118 - 1 UUAUUUUAUUUGCCAUAUGUUGC-GAGGCACUGGAAAUGUACUAGUUAUGGCCUGACCAUAUGCUCAGCGGAGACGCACAC-CACGGAGCAGUACUUUGCUAGGCCUUUCGCGGAGCAGG .........((((......((((-(((((.((((....(((((...(((((.....))))).((((.(((....))).(..-...))))))))))....)))))))..)))))).)))). ( -39.20) >DroYak_CAF1 9652 99 - 1 UUAUU----AUGCCAUAUGUUGC-GAGGCACUGAAAAUGUACUAGUUAUAGCCCCACAAUAUGCGCAGCGGAGACGCACAC-CGCGGUGCAUUACCUUGCUACAC--------------- .....----........(((.((-((((.......((((((((.((....))..........(((..(((....)))....-))))))))))).)))))).))).--------------- ( -25.40) >consensus UUAUU____GUGCCAUAUGUUGCGGUGGCACUGAAAAUGUACUAAAUAUAGCCAUACAAUACGCACAGCGGAGACGCACAC_CAGGGUGCAUGACUCUGCUA__C__________A___A .........(((((((........))))))).....(((((((.......((..........))...(((....)))........)))))))............................ (-17.60 = -18.44 + 0.84)

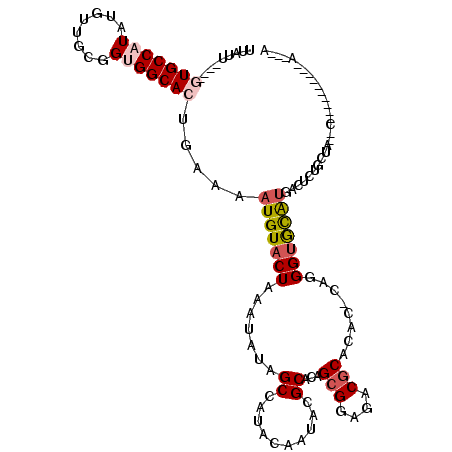
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:45 2006