
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,958,144 – 7,958,259 |
| Length | 115 |
| Max. P | 0.993642 |

| Location | 7,958,144 – 7,958,238 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7958144 94 + 22407834 GUUA-------------UUCGAUCCCGAACUCAUUUCUUGCCCUGCA---CCAACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGC ....-------------((((....))))...............(((---.....((((((((((...........))))))))))..((((((......)))))).))) ( -21.10) >DroSec_CAF1 129466 91 + 1 GUUG-------------UUCGAUCCCGAACUCAUUUCUUGCCAUGC------AACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGC ...(-------------((((....)))))..............((------(..((((((((((...........))))))))))..((((((......)))))).))) ( -24.50) >DroSim_CAF1 129535 97 + 1 GUUG-------------UUCGAUCCCGAACUAAUUUCUUGCCAUGCACCACCAACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGC ...(-------------((((....)))))..............(((........((((((((((...........))))))))))..((((((......)))))).))) ( -24.20) >DroEre_CAF1 131324 107 + 1 GUUUUAAACAGAUCUCUGUCGAUGCCGAACUCAUUUCUUGCCAUGCA---CCAACUGGCAACAUCAAUCUGGGCAAGAUGUUGCCACAUUAAAUUAAAAUAUUCAAAUGC (((((((((((....)))).((((..............(((...)))---.....((((((((((...........))))))))))))))...))))))).......... ( -21.20) >DroYak_CAF1 134693 105 + 1 GUUUUAAACAGAUCU--UUCGAUCGUGAACUCAUUUCUCGCCAUGCA---CCAACUGGCAACAUCAAUGUGGGCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGC ..........((((.--...))))((((.........))))...(((---.....((((((((((..((....)).))))))))))..((((((......)))))).))) ( -24.90) >consensus GUUG_____________UUCGAUCCCGAACUCAUUUCUUGCCAUGCA___CCAACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGC ..................(((....)))................((.........((((((((((...........))))))))))..((((((......))))))..)) (-17.98 = -18.02 + 0.04)


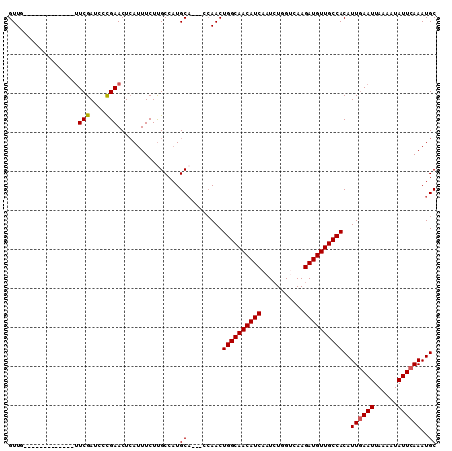
| Location | 7,958,144 – 7,958,238 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7958144 94 - 22407834 GCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUUGG---UGCAGGGCAAGAAAUGAGUUCGGGAUCGAA-------------UAAC (((((.((((......)))))))))(((((((((...........)))))))))..((((---(.(.((((.(....)..)))).).))))).-------------.... ( -26.10) >DroSec_CAF1 129466 91 - 1 GCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUU------GCAUGGCAAGAAAUGAGUUCGGGAUCGAA-------------CAAC (((.((((((......))))))..((((((((((...........))))))))))..)------))..............(((((....))))-------------)... ( -26.60) >DroSim_CAF1 129535 97 - 1 GCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUUGGUGGUGCAUGGCAAGAAAUUAGUUCGGGAUCGAA-------------CAAC ((((((((((......))))))..((((((((((...........)))))))))).......))))..............(((((....))))-------------)... ( -27.70) >DroEre_CAF1 131324 107 - 1 GCAUUUGAAUAUUUUAAUUUAAUGUGGCAACAUCUUGCCCAGAUUGAUGUUGCCAGUUGG---UGCAUGGCAAGAAAUGAGUUCGGCAUCGACAGAGAUCUGUUUAAAAC ...((((((((..((.........((((((((((...........))))))))))(((((---(((..(((.(....)..)))..))))))))...))..)))))))).. ( -31.50) >DroYak_CAF1 134693 105 - 1 GCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGCCCACAUUGAUGUUGCCAGUUGG---UGCAUGGCGAGAAAUGAGUUCACGAUCGAA--AGAUCUGUUUAAAAC ...((((((((.......((((((((((((....))).)))))))))(.((((((((...---.)).)))))).)...........((((...--.)))))))))))).. ( -32.40) >consensus GCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUUGG___UGCAUGGCAAGAAAUGAGUUCGGGAUCGAA_____________AAAC ((..((((((......))))))..((((((((((...........)))))))))).........))............................................ (-20.22 = -20.06 + -0.16)

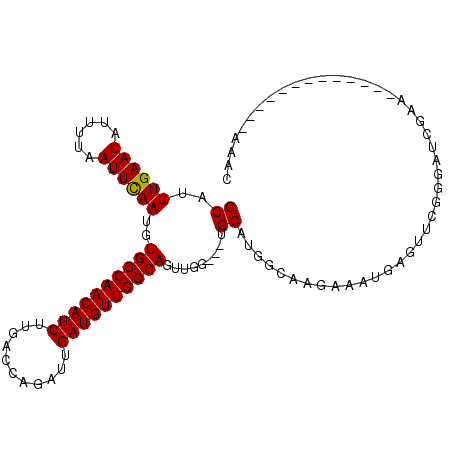
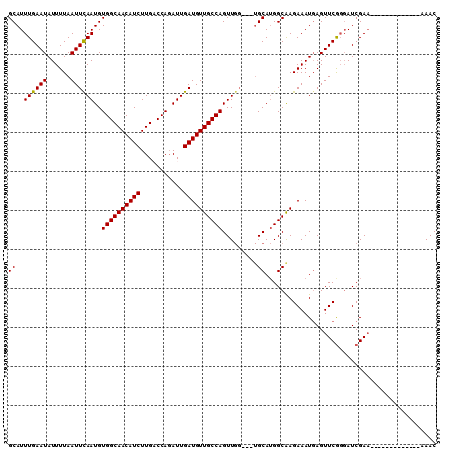
| Location | 7,958,169 – 7,958,259 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.86 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7958169 90 + 22407834 UGCCCUGCA---CCAACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGCAACCA--------AGUGGAUG-GGAGUGGG ...((..(.---((...((((((((((...........))))))))))..((((((......))))))...((.((.--------...)).))-)).)..)) ( -27.60) >DroSec_CAF1 129491 96 + 1 UGCCAUGC------AACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGCAACCAUCCAGCCAAGUGGAUGGGGAGUGGG ..((((((------(..((((((((((...........))))))))))..((((((......)))))).)))..(((((((......)))))))...)))). ( -36.30) >DroSim_CAF1 129560 92 + 1 UGCCAUGCACCACCAACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGCAACCAUCC----------AUGGGGAGUGGG ..((((...((.(((..((((((((((...........))))))))))..((((((......))))))............----------.))))).)))). ( -28.20) >DroEre_CAF1 131362 94 + 1 UGCCAUGCA---CCAACUGGCAACAUCAAUCUGGGCAAGAUGUUGCCACAUUAAAUUAAAAUAUUCAAAUGCAAGCAGCC----AAGAAGCUG-GGAGUGGG ..((((...---((...((((((((((...........))))))))))...........................((((.----.....))))-)).)))). ( -25.70) >DroYak_CAF1 134729 94 + 1 CGCCAUGCA---CCAACUGGCAACAUCAAUGUGGGCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGCAGCCAGCC----AAGAAGAUG-GGAGUGGG ..((((...---(((.(((((((((((..((....)).)))))))))...((((((......))))))............----....)).))-)..)))). ( -27.90) >consensus UGCCAUGCA___CCAACUGGCAACAUCAAUCUGGUCAAGAUGUUGCCACAUUGAAUUAAAAUAUUCAAAUGCAACCAGCC____AAGAAGAUG_GGAGUGGG ..((((((.........((((((((((...........))))))))))..((((((......))))))..))...((((..........))))....)))). (-20.82 = -21.86 + 1.04)


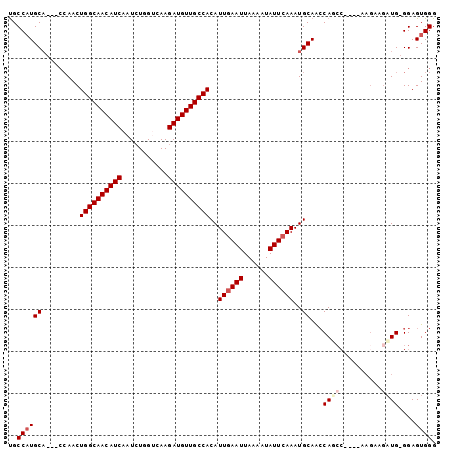
| Location | 7,958,169 – 7,958,259 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7958169 90 - 22407834 CCCACUCC-CAUCCACU--------UGGUUGCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUUGG---UGCAGGGCA (((....(-((......--------))).(((((((((((......))))))..((((((((((...........))))))))))....)---))))))).. ( -29.50) >DroSec_CAF1 129491 96 - 1 CCCACUCCCCAUCCACUUGGCUGGAUGGUUGCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUU------GCAUGGCA .(((....(((((((......))))))).((((.((((((......))))))..((((((((((...........))))))))))..)------)))))).. ( -33.70) >DroSim_CAF1 129560 92 - 1 CCCACUCCCCAU----------GGAUGGUUGCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUUGGUGGUGCAUGGCA .(((..(.((((----------.(((....(((((.((((......)))))))))(((((((((...........))))))))).))).)))).)..))).. ( -29.10) >DroEre_CAF1 131362 94 - 1 CCCACUCC-CAGCUUCUU----GGCUGCUUGCAUUUGAAUAUUUUAAUUUAAUGUGGCAACAUCUUGCCCAGAUUGAUGUUGCCAGUUGG---UGCAUGGCA .(((....-(((((....----)))))..(((((((((((......))))))..((((((((((...........))))))))))....)---))))))).. ( -28.20) >DroYak_CAF1 134729 94 - 1 CCCACUCC-CAUCUUCUU----GGCUGGCUGCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGCCCACAUUGAUGUUGCCAGUUGG---UGCAUGGCG .......(-((((...(.----.((((((.(((((.((((......))))((((((((((....))).)))))))))))).))))))..)---.).)))).. ( -31.10) >consensus CCCACUCC_CAUCCACUU____GGAUGGUUGCAUUUGAAUAUUUUAAUUCAAUGUGGCAACAUCUUGACCAGAUUGAUGUUGCCAGUUGG___UGCAUGGCA .((......(((((........)))))..(((..((((((......))))))..((((((((((...........)))))))))).........))).)).. (-22.70 = -23.50 + 0.80)

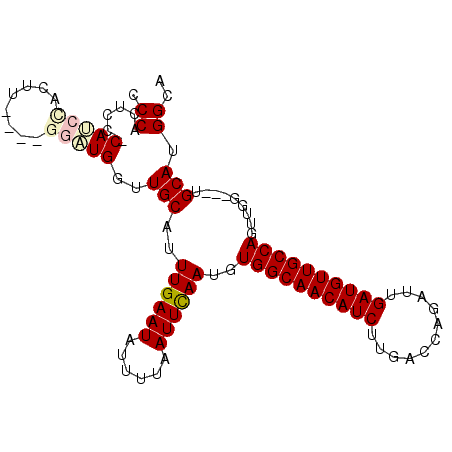
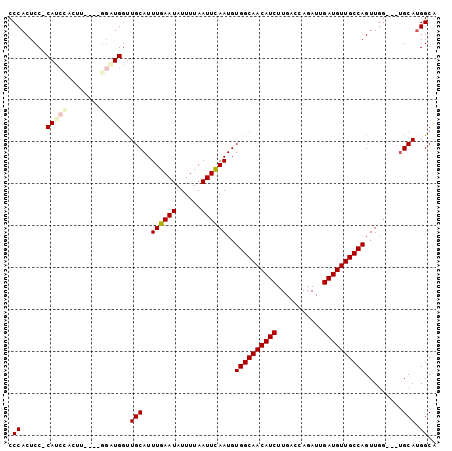
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:38 2006