
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,957,960 – 7,958,065 |
| Length | 105 |
| Max. P | 0.966504 |

| Location | 7,957,960 – 7,958,065 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.11 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -8.46 |
| Energy contribution | -13.46 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
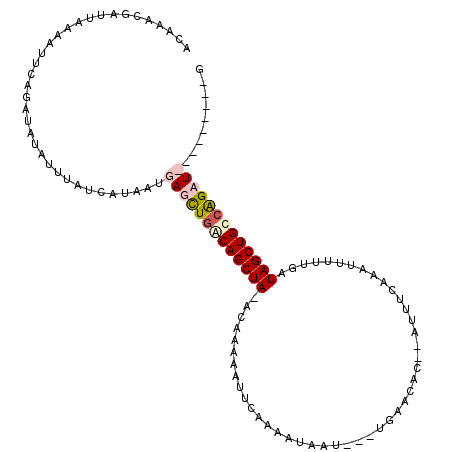
>2L_DroMel_CAF1 7957960 105 + 22407834 C---------AUCUGACAGCUAUCAAAAAUUUGAAAU--GUGUUCAUACAUUAUUUGGAAAUUUUGUUUAGCUGCAAGCUCAUUAUGAUAAAUAGAUACCAUUUAUAAUCGUUUGU .---------......((((((.(((((.(((.((((--((((....))).))))).))).)))))..))))))(((((..(((((((..............))))))).))))). ( -21.74) >DroSim_CAF1 129388 81 + 1 C---------AUCUGACAGCUAUC-------------------------AUUAUUUGGAAUUUUUGU-UAGCUGUCAGCUCAUUAUGAUAAAUAGAUACCAAUUAUAAUCGUUUGA .---------..((((((((((.(-------------------------(..(........)..)).-))))))))))...((((((((............))))))))....... ( -17.60) >DroEre_CAF1 131129 108 + 1 UAUCUUAGUAAUCCGGCAGCUAUCGAAAAUUUGAAAUUUGUGUUCA-------UUUUGAAUUUUUGU-UAGCUGUCAGAUCAAUAUGAUAAAUAUAUCUGAACAUUAAUCGUUUGU ((((.((...(((.((((((((.((((((((..((((........)-------)))..)))))))).-)))))))).)))...)).)))).........((((.......)))).. ( -28.20) >DroYak_CAF1 134496 113 + 1 CAACUUAGUAAUCUGGCAGCUAUUAAAAAUUGAAAAU--GUGUUCAUACAUUAUAUUGAAUUUUUGU-UAGCUGUCAAAUCGUAAUGAUAAAAAUAACUGAAUAUUAAUCGUUUGC ....((((((.(((((((((((.((((((((.(((((--(((....))))))...)).)))))))).-))))))))).(((.....)))..........)).))))))........ ( -24.00) >consensus C_________AUCUGACAGCUAUCAAAAAUUUGAAAU__GUGUUCA___AUUAUUUGGAAUUUUUGU_UAGCUGUCAGAUCAUUAUGAUAAAUAGAUACCAAUAAUAAUCGUUUGU ............(((((((((..((((((((((((((...............))))))))))))))...)))))))))...................................... ( -8.46 = -13.46 + 5.00)


| Location | 7,957,960 – 7,958,065 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.11 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -8.61 |
| Energy contribution | -9.93 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7957960 105 - 22407834 ACAAACGAUUAUAAAUGGUAUCUAUUUAUCAUAAUGAGCUUGCAGCUAAACAAAAUUUCCAAAUAAUGUAUGAACAC--AUUUCAAAUUUUUGAUAGCUGUCAGAU---------G ..............((((((......))))))......((.(((((((...(((((((......(((((......))--)))..)))))))...))))))).))..---------. ( -17.20) >DroSim_CAF1 129388 81 - 1 UCAAACGAUUAUAAUUGGUAUCUAUUUAUCAUAAUGAGCUGACAGCUA-ACAAAAAUUCCAAAUAAU-------------------------GAUAGCUGUCAGAU---------G ........((((...(((((......)))))..)))).((((((((((-.((..............)-------------------------).))))))))))..---------. ( -18.54) >DroEre_CAF1 131129 108 - 1 ACAAACGAUUAAUGUUCAGAUAUAUUUAUCAUAUUGAUCUGACAGCUA-ACAAAAAUUCAAAA-------UGAACACAAAUUUCAAAUUUUCGAUAGCUGCCGGAUUACUAAGAUA ......(((.(((((......))))).)))....(((((((.((((((-.(.((((((..(((-------(........))))..)))))).).)))))).)))))))........ ( -22.90) >DroYak_CAF1 134496 113 - 1 GCAAACGAUUAAUAUUCAGUUAUUUUUAUCAUUACGAUUUGACAGCUA-ACAAAAAUUCAAUAUAAUGUAUGAACAC--AUUUUCAAUUUUUAAUAGCUGCCAGAUUACUAAGUUG ...((((((.((((......))))...))).....((((((.((((((-..(((((((......(((((......))--)))...)))))))..)))))).)))))).....))). ( -19.60) >consensus ACAAACGAUUAAAAUUCAGAUAUAUUUAUCAUAAUGAGCUGACAGCUA_ACAAAAAUUCAAAAUAAU___UGAACAC__AUUUCAAAUUUUUGAUAGCUGCCAGAU_________G ...................................(((((((((((((..............................................)))))))))))))......... ( -8.61 = -9.93 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:35 2006