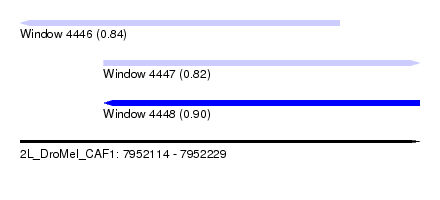
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,952,114 – 7,952,229 |
| Length | 115 |
| Max. P | 0.903077 |

| Location | 7,952,114 – 7,952,206 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -18.33 |
| Consensus MFE | -13.72 |
| Energy contribution | -12.83 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7952114 92 - 22407834 CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAUUGCCAACGGCCAUGCAGUUUUCCACUCA-CCCGCCGCCC---------CUUAC .(((((((((..((((.(((((....))))).))))...))).))))))......((((..((.(((.....)))))-...))))...---------..... ( -18.40) >DroPse_CAF1 151963 100 - 1 CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAUUGCCACUGGCCACACCACUGGCCCUUC--CCCUUCACCUAGCUCCCCACCAUC .(((((((((..((((.(((((....))))).))))...))).)))))).((((.(((.....))).)))).....--........................ ( -20.70) >DroSec_CAF1 123650 90 - 1 CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCUGUCUGCAAAUUGCCAACGGCCAUGCAGUUUUCACCGC--CCCUCC-CCG---------CACCC .((((((((((.((((.(((((....))))).))))..)))).))))))(((....((....((.((....)).))--....))-..)---------))... ( -17.20) >DroSim_CAF1 123691 91 - 1 CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAUUGCCAACGGCCAUGCAGUUUUCCCCGC--CCCGCCCCCU---------CCCUC (((((((.......)))))))...((((((......(((((..((.....))..))))).))))))..........--..........---------..... ( -16.70) >DroAna_CAF1 122055 91 - 1 CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAUUGCCACUGGCCACAGAGUUCUCCCUUCAACCUGCCACCU---------CCA-- .(((((((((..((((.(((((....))))).))))...))).))))))......((((...((.(((........)))))))))...---------...-- ( -16.30) >DroPer_CAF1 151428 100 - 1 CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAUUGCCACUGGCCACACCACUGGCCCUUC--CCCUUCACCUAGCUCCCCACCAUC .(((((((((..((((.(((((....))))).))))...))).)))))).((((.(((.....))).)))).....--........................ ( -20.70) >consensus CAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAUUGCCAACGGCCACACAGUUUUCCCCUC__CCCGCCACCU_________CCAUC ....((((....((((.(((((....))))).))))((((...((.....))...))))))))....................................... (-13.72 = -12.83 + -0.89)
| Location | 7,952,138 – 7,952,229 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
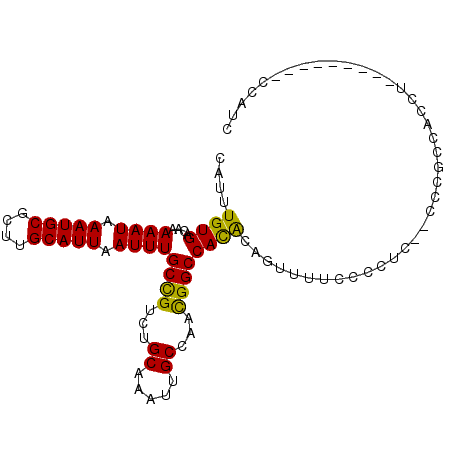
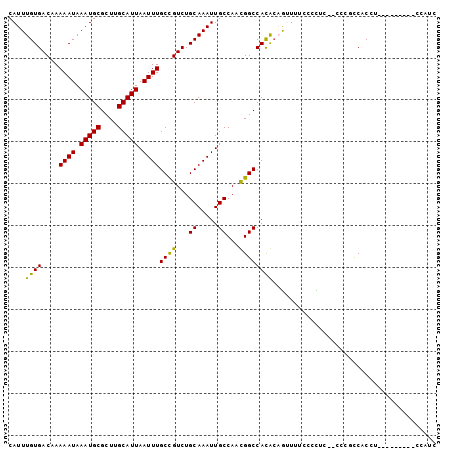
>2L_DroMel_CAF1 7952138 91 + 22407834 AACUG------CAUGGCCGUUGGCA--AUUUGCAGACGGCAAAUUAAUGCAAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACGGCA---GCAAAAGCAAC-- ...((------(...(((((((...--.((((((...((((((..(((((....)))))....)))))).....))))))))))))).---)))........-- ( -29.30) >DroVir_CAF1 136521 92 + 1 GGUAGGAGGCCGCUGGCCACUGGCAA-AAUUGCAGACGGCAAAUUAAUGCCAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACAGCA---ACAA---C----- (((.....)))(((((((...)))..-..(((((...((((((..(((((....)))))....)))))).....)))))....)))).---....---.----- ( -25.90) >DroSec_CAF1 123672 91 + 1 AACUG------CAUGGCCGUUGGCA--AUUUGCAGACAGCAAAUUAAUGCAAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACAGCA---GCUAAAGCAAC-- ...((------(.((((.((((...--.(((((((((((.((((.(((((....))))).))))))))).....))))))...)))).---))))..)))..-- ( -25.90) >DroSim_CAF1 123714 91 + 1 AACUG------CAUGGCCGUUGGCA--AUUUGCAGACGGCAAAUUAAUGCAAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACAGCA---GCUAAAGCAAC-- ...((------(.((((.((((...--.((((((...((((((..(((((....)))))....)))))).....))))))...)))).---))))..)))..-- ( -26.50) >DroMoj_CAF1 150900 96 + 1 GUUUGGUGCCCGCUGGCCACUGGCAAAAAUUGCAGACGGCAAAUUAAUGCCAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACAGCA---ACAAGAGC----- ((((((((((....)).)))(((((((((((((.(..((((......))))..))))...))))))))))......))))).......---........----- ( -24.40) >DroAna_CAF1 122078 96 + 1 AACUC------UGUGGCCAGUGGCA--AUUUGCAGACGGCAAAUUAAUGCAAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACAGCAACGGCAACAGCAACGG ....(------(((.(((.((((((--((((((.....)))))))(((((....))))).......)))))...(((........)))..))).))))...... ( -28.00) >consensus AACUG______CAUGGCCAUUGGCA__AUUUGCAGACGGCAAAUUAAUGCAAGCGCAUUUAUUUUUGUCACAAAUGCAAACAACAGCA___GCAAAAGCAAC__ ...............(((...))).....(((((...((((((..(((((....)))))....)))))).....)))))......................... (-16.37 = -16.53 + 0.17)

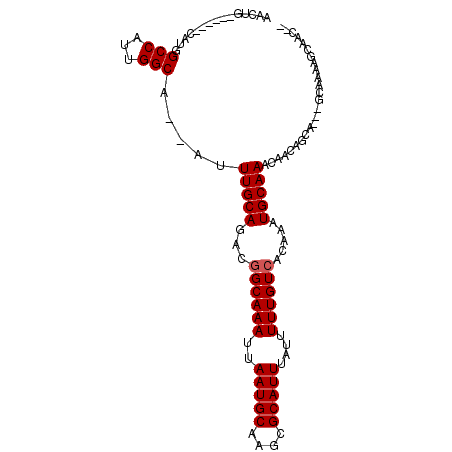
| Location | 7,952,138 – 7,952,229 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.95 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7952138 91 - 22407834 --GUUGCUUUUGC---UGCCGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAU--UGCCAACGGCCAUG------CAGUU --.......((((---.(((((((...(((((((((((..((((.(((((....))))).))))...))).))))).--))))))))))...)------))).. ( -29.20) >DroVir_CAF1 136521 92 - 1 -----G---UUGU---UGCUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUGGCAUUAAUUUGCCGUCUGCAAUU-UUGCCAGUGGCCAGCGGCCUCCUACC -----.---....---.(((((((...((((((((.......))))))))(((((((.....((((.....))))..-.)))))))...)))))))........ ( -31.50) >DroSec_CAF1 123672 91 - 1 --GUUGCUUUAGC---UGCUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCUGUCUGCAAAU--UGCCAACGGCCAUG------CAGUU --........(((---(((.((((((.((((((((((((.((((.(((((....))))).))))..)))).))))).--))).))))))...)------))))) ( -30.40) >DroSim_CAF1 123714 91 - 1 --GUUGCUUUAGC---UGCUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAU--UGCCAACGGCCAUG------CAGUU --........(((---(((.((((((.(((((((((((..((((.(((((....))))).))))...))).))))).--))).))))))...)------))))) ( -28.50) >DroMoj_CAF1 150900 96 - 1 -----GCUCUUGU---UGCUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUGGCAUUAAUUUGCCGUCUGCAAUUUUUGCCAGUGGCCAGCGGGCACCAAAC -----....(((.---((((((((...((((((((.......))))))))(((((((..((((.((.....))))))..)))))))...)))).)))).))).. ( -32.00) >DroAna_CAF1 122078 96 - 1 CCGUUGCUGUUGCCGUUGCUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAU--UGCCACUGGCCACA------GAGUU ......((((.(((((.((...(((.(((((((((.......)))))))))...)))..(((((((.....))))))--))).)).))).)))------).... ( -27.10) >consensus __GUUGCUUUUGC___UGCUGUUGUUUGCAUUUGUGACAAAAAUAAAUGCGCUUGCAUUAAUUUGCCGUCUGCAAAU__UGCCAACGGCCAUG______CAGUU .................(((((((...((((((((.......))))))))....(((...((((((.....))))))..))))))))))............... (-22.00 = -21.95 + -0.05)

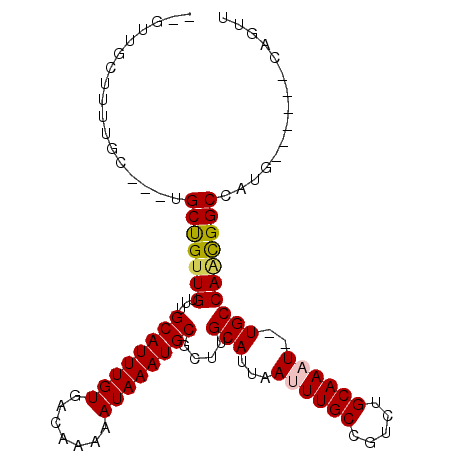

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:30 2006