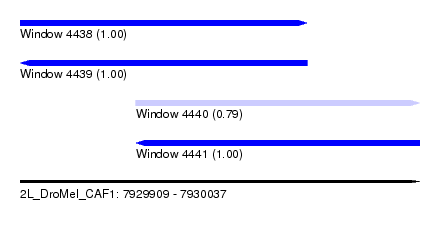
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,929,909 – 7,930,037 |
| Length | 128 |
| Max. P | 0.999722 |

| Location | 7,929,909 – 7,930,001 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -28.08 |
| Energy contribution | -28.56 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7929909 92 + 22407834 GAGUCGCGUCCGUCCGUGUCAUUGUCAUAGCAAGCAAACAAGCGGAGUAGCUGGCUUUGCCUAGCAACUGCCACUUGGAGUGCCACUCAAAA ((((.((((((....((....((((....))))....))(((.(((((.((((((...))).))).))).)).)))))).))).)))).... ( -29.10) >DroSec_CAF1 101872 88 + 1 GAGUCGCGU----CCGUGUCAUUGUCAUAGCAAACAAACAAGCGGAGUAGCUGGCUUUGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAA ((((.(((.----.(.(((..((((....)))))))..((((.((((((((((((...))).))))))).)).)))))..))).)))).... ( -31.40) >DroSim_CAF1 101840 88 + 1 GAGUCGCGU----CCGUGUCAUUGUCAUAGCAAGCAAACAAGCGGAGUAGCUGGCUUUGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAA ((((.(((.----.(.(((..((((....)))))))..((((.((((((((((((...))).))))))).)).)))))..))).)))).... ( -31.70) >DroEre_CAF1 103293 87 + 1 GAGUCGCGU----CCGUGCCAUUGUCAAAGCA-GCAAGCAAGCGGAGUAGCUGGCUUAGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAA ((((.(((.----.(.(((...(((....)))-)))..((((.((((((((((((...))).))))))).)).)))))..))).)))).... ( -31.50) >DroYak_CAF1 105264 88 + 1 GAGUCGCGU----CCGUGUCAUUGUCAUAGCAAGCAAACAAGCGGAGUAGCUGGCUUUGCAUUGCUACUGCCACUUGUUCUGCCACUCAAAA ((((.((..----...(((..((((....)))))))((((((.((((((((..((...))...)))))).)).))))))..)).)))).... ( -27.90) >consensus GAGUCGCGU____CCGUGUCAUUGUCAUAGCAAGCAAACAAGCGGAGUAGCUGGCUUUGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAA ((((.((.........(((..((((....)))))))..((((.((((((((((((...))).))))))).)).))))....)).)))).... (-28.08 = -28.56 + 0.48)

| Location | 7,929,909 – 7,930,001 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -29.22 |
| Energy contribution | -30.46 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7929909 92 - 22407834 UUUUGAGUGGCACUCCAAGUGGCAGUUGCUAGGCAAAGCCAGCUACUCCGCUUGUUUGCUUGCUAUGACAAUGACACGGACGGACGCGACUC ....((((.((..(((((((((.(((.(((.(((...)))))).)))))))))(((((..((.(((....))).)))))))))).)).)))) ( -31.20) >DroSec_CAF1 101872 88 - 1 UUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCAAAGCCAGCUACUCCGCUUGUUUGUUUGCUAUGACAAUGACACGG----ACGCGACUC ..(((.((((((((((((((((.(((((((.(((...)))))))))))))))))...)))))))))..)))........----......... ( -35.80) >DroSim_CAF1 101840 88 - 1 UUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCAAAGCCAGCUACUCCGCUUGUUUGCUUGCUAUGACAAUGACACGG----ACGCGACUC ....(((((((((((.((((((.(((((((.(((...)))))))))))))))))))))))................((.----...)))))) ( -33.60) >DroEre_CAF1 103293 87 - 1 UUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCUAAGCCAGCUACUCCGCUUGCUUGC-UGCUUUGACAAUGGCACGG----ACGCGACUC ....((((.((....(((((((.(((((((.(((...))))))))))))))))).(((.-((((........)))))))----..)).)))) ( -36.10) >DroYak_CAF1 105264 88 - 1 UUUUGAGUGGCAGAACAAGUGGCAGUAGCAAUGCAAAGCCAGCUACUCCGCUUGUUUGCUUGCUAUGACAAUGACACGG----ACGCGACUC ....((((.((.((((((((((.((((((...((...))..)))))))))))))))).....(..((.......))..)----..)).)))) ( -30.30) >consensus UUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCAAAGCCAGCUACUCCGCUUGUUUGCUUGCUAUGACAAUGACACGG____ACGCGACUC ....((((((((((.(((((((.(((((((.(((...)))))))))))))))))))))))..............(.((......)).))))) (-29.22 = -30.46 + 1.24)

| Location | 7,929,946 – 7,930,037 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7929946 91 + 22407834 ACAAGCGGAGUAGCUGGCUUUGCCUAGCAACUGCCACUUGGAGUGCCACUCAAAAUCAGAUCCCGACUUGAGACCAGUGAUCCG--GGGAAGA .((((.(((((.((((((...))).))).))).)).))))(((.....))).........(((((.................))--))).... ( -25.23) >DroSec_CAF1 101905 92 + 1 ACAAGCGGAGUAGCUGGCUUUGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAAUCAGAACCCGACUUGAGACCAGAGUUUCGGG-GGGAGA .((((.((((((((((((...))).))))))).)).)))).((((...........)))).(((..((((((((....))))))))-)))... ( -34.00) >DroSim_CAF1 101873 93 + 1 ACAAGCGGAGUAGCUGGCUUUGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAAUCAGAACCCGACUUGAGACCAGAGUUUCUGGGGGAAGA .((((.((((((((((((...))).))))))).)).)))).(((.((.(((((..((.......)).))))).((((.....)))).)).))) ( -34.30) >DroEre_CAF1 103325 91 + 1 GCAAGCGGAGUAGCUGGCUUAGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAAUCAGAUCCCGACUUGAGACCAGAGUUCCG--GGGGACA .((((.((((((((((((...))).))))))).)).))))((((.((((((....((((........)))).....))))...)--).)))). ( -34.60) >DroYak_CAF1 105297 91 + 1 ACAAGCGGAGUAGCUGGCUUUGCAUUGCUACUGCCACUUGUUCUGCCACUCAAAAUCAGAUCCCGACUUGAGACCAGAGUUCCG--GAGCCCU (((((.((((((((..((...))...)))))).)).)))))((((..((((....((((........)))).....))))..))--))..... ( -26.20) >consensus ACAAGCGGAGUAGCUGGCUUUGCCUAGCUACUGCCACUUGGUCUGCCACUCAAAAUCAGAUCCCGACUUGAGACCAGAGUUCCG__GGGAAGA .((((.((((((((((((...))).))))))).)).)))).((((...(((((..((.......)).)))))..))))............... (-24.46 = -24.94 + 0.48)


| Location | 7,929,946 – 7,930,037 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -32.06 |
| Energy contribution | -32.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7929946 91 - 22407834 UCUUCCC--CGGAUCACUGGUCUCAAGUCGGGAUCUGAUUUUGAGUGGCACUCCAAGUGGCAGUUGCUAGGCAAAGCCAGCUACUCCGCUUGU .......--((((((.((((.(....))))))))))).....(((.....)))(((((((.(((.(((.(((...)))))).)))))))))). ( -31.50) >DroSec_CAF1 101905 92 - 1 UCUCCC-CCCGAAACUCUGGUCUCAAGUCGGGUUCUGAUUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCAAAGCCAGCUACUCCGCUUGU ......-........((((.(((((((((((...)))).)))))).).)))).(((((((.(((((((.(((...))))))))))))))))). ( -35.60) >DroSim_CAF1 101873 93 - 1 UCUUCCCCCAGAAACUCUGGUCUCAAGUCGGGUUCUGAUUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCAAAGCCAGCUACUCCGCUUGU (((.((.((((.....)))).((((((((((...)))).)))))).)).))).(((((((.(((((((.(((...))))))))))))))))). ( -38.70) >DroEre_CAF1 103325 91 - 1 UGUCCCC--CGGAACUCUGGUCUCAAGUCGGGAUCUGAUUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCUAAGCCAGCUACUCCGCUUGC ((((.(.--(((((.((.((((((.....)))))).))))))).).))))...(((((((.(((((((.(((...))))))))))))))))). ( -37.90) >DroYak_CAF1 105297 91 - 1 AGGGCUC--CGGAACUCUGGUCUCAAGUCGGGAUCUGAUUUUGAGUGGCAGAACAAGUGGCAGUAGCAAUGCAAAGCCAGCUACUCCGCUUGU ...((((--(((....)))).((((((((((...)))).)))))).)))...((((((((.((((((...((...))..)))))))))))))) ( -32.40) >consensus UCUCCCC__CGGAACUCUGGUCUCAAGUCGGGAUCUGAUUUUGAGUGGCAGACCAAGUGGCAGUAGCUAGGCAAAGCCAGCUACUCCGCUUGU ...............((((.(((((((((((...)))).)))))).).)))).(((((((.(((((((.(((...))))))))))))))))). (-32.06 = -32.54 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:24 2006