
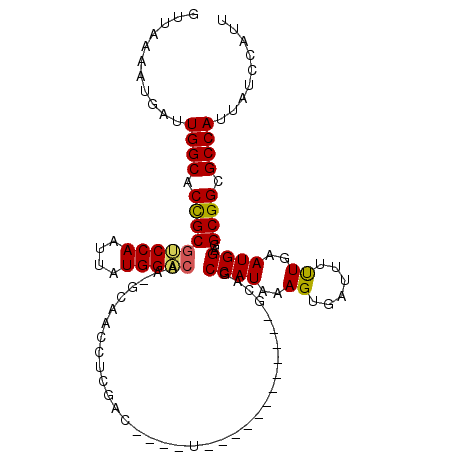
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,897,618 – 7,897,722 |
| Length | 104 |
| Max. P | 0.975671 |

| Location | 7,897,618 – 7,897,722 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7897618 104 + 22407834 GUUAAAAUGAUUGGCACUGCGUCCAAUUAUGGACAAGGCAACCUCGGC----UGCAUUC---GCAGCAGCCAUAAAGUGAUUUUUGAAUGGAGGCGGCGCCAUUAUCCAUU ......(((((.(((.((..(((((....))))).))((..(((((((----(((....---...)))))).(((((....)))))....))))..))))))))))..... ( -36.10) >DroVir_CAF1 66246 90 + 1 GUUAAAAUGAUUGGCACCGCGUCCAAUUAUGGCCC---CAACGACGAC----U-------------GAGCCAUAAAGUGAUU-CUGAAUGGAGGCGGCGCCAGCUGCCAAC ..........(((((.((((.((((.(((((((..---((........----)-------------).)))))))((.....-))...)))).)))).)))))........ ( -30.50) >DroEre_CAF1 70420 94 + 1 GUUAAAAUGAUUGGCACUGCGUCCAAUUAUGGACAA-GCAACCUCAGC----------------GGCAGCCAUAAAGUGAUUUUUGAAUGGAGGCGGCGCCAUUAUCCAUU ......(((((.(((..((((((((....)))))..-)))......((----------------.((..((((..((......))..))))..)).))))))))))..... ( -26.50) >DroMoj_CAF1 74215 87 + 1 GUUAAAAUGAUUGGCACCGCGGCCAAUUAUGGCCC---C---GUCGAC----U-------------CAGCCAUAAAGUGAUU-UUGAAUGGAGGCGGCGCCAGUUGCCAGU ........(((((((.(((((((((....)))))(---(---(((((.----.-------------..((......))....-))).))))..)))).)))))))...... ( -31.30) >DroAna_CAF1 69889 103 + 1 GUUAAAAUGAUUGGCACUGCUUCCAAUUAUGGACAACGCAACCGCG--------CAGUUGCUGCAGACGCCAUAAAGUGAUUUUUGAAUGGAGGCGGCGCCAUUAUCCAUU ......(((((.(((.(((((((((.(((..((..(((((((....--------..))))).((....))......))..))..))).))))))))).))))))))..... ( -32.20) >DroPer_CAF1 74847 107 + 1 GUUAAAAUGAUUGGCACCGCGUCCAAUUAUGGACAAGGAAACCCGAACGAACCUCAGAC---GC-GCAGCCAUAAAGUGAUUUUUGAAUGGAGGCGGCGCCAUUAUCCAUU ......(((((.(((.((((.((((....(((....(....))))........(((((.---.(-((.........)))...))))).)))).)))).))))))))..... ( -29.10) >consensus GUUAAAAUGAUUGGCACCGCGUCCAAUUAUGGACAA_GCAACCUCGAC____U____________GCAGCCAUAAAGUGAUUUUUGAAUGGAGGCGGCGCCAUUAUCCAUU ...........((((.(((((((((....)))))...................................((((..((......))..))))..)))).))))......... (-22.46 = -22.18 + -0.28)


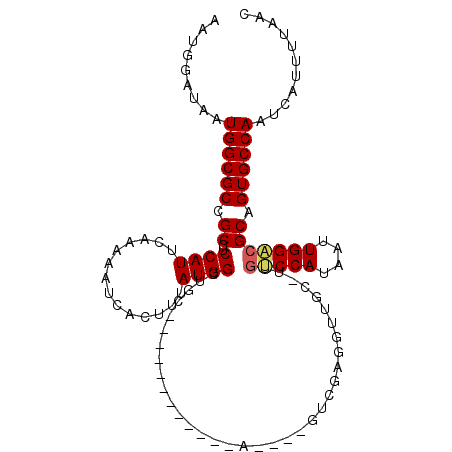
| Location | 7,897,618 – 7,897,722 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.71 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7897618 104 - 22407834 AAUGGAUAAUGGCGCCGCCUCCAUUCAAAAAUCACUUUAUGGCUGCUGC---GAAUGCA----GCCGAGGUUGCCUUGUCCAUAAUUGGACGCAGUGCCAAUCAUUUUAAC ....(((..((((((.(((((.....(((......)))..((((((...---....)))----))))))))(((...(((((....)))))))))))))))))........ ( -35.00) >DroVir_CAF1 66246 90 - 1 GUUGGCAGCUGGCGCCGCCUCCAUUCAG-AAUCACUUUAUGGCUC-------------A----GUCGUCGUUG---GGGCCAUAAUUGGACGCGGUGCCAAUCAUUUUAAC .........(((((((((.((((.....-.......(((((((((-------------.----..........---))))))))).)))).)))))))))........... ( -33.62) >DroEre_CAF1 70420 94 - 1 AAUGGAUAAUGGCGCCGCCUCCAUUCAAAAAUCACUUUAUGGCUGCC----------------GCUGAGGUUGC-UUGUCCAUAAUUGGACGCAGUGCCAAUCAUUUUAAC ....(((..((((((.((......................(((.(((----------------.....))).))-).(((((....))))))).)))))))))........ ( -27.40) >DroMoj_CAF1 74215 87 - 1 ACUGGCAACUGGCGCCGCCUCCAUUCAA-AAUCACUUUAUGGCUG-------------A----GUCGAC---G---GGGCCAUAAUUGGCCGCGGUGCCAAUCAUUUUAAC ...(....)(((((((((..(((.....-.......((((((((.-------------.----(....)---.---.)))))))).)))..)))))))))........... ( -31.02) >DroAna_CAF1 69889 103 - 1 AAUGGAUAAUGGCGCCGCCUCCAUUCAAAAAUCACUUUAUGGCGUCUGCAGCAACUG--------CGCGGUUGCGUUGUCCAUAAUUGGAAGCAGUGCCAAUCAUUUUAAC ((((((....(((...)))))))))..............(((((.((((.((((((.--------...))))))....((((....)))).)))))))))........... ( -32.70) >DroPer_CAF1 74847 107 - 1 AAUGGAUAAUGGCGCCGCCUCCAUUCAAAAAUCACUUUAUGGCUGC-GC---GUCUGAGGUUCGUUCGGGUUUCCUUGUCCAUAAUUGGACGCGGUGCCAAUCAUUUUAAC ....(((..(((((((((......................((..((-.(---(..((.....))..)).))..))..(((((....)))))))))))))))))........ ( -34.00) >consensus AAUGGAUAAUGGCGCCGCCUCCAUUCAAAAAUCACUUUAUGGCUGC____________A____GUCGAGGUUGC_UUGUCCAUAAUUGGACGCAGUGCCAAUCAUUUUAAC .........((((((.((..((((..............))))...................................(((((....))))))).))))))........... (-19.59 = -19.71 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:15 2006