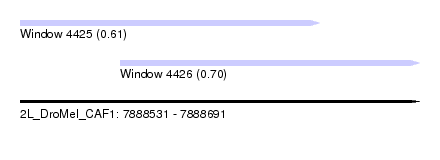
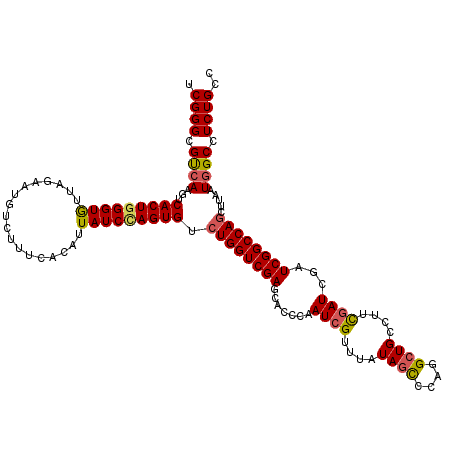
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,888,531 – 7,888,691 |
| Length | 160 |
| Max. P | 0.702639 |

| Location | 7,888,531 – 7,888,651 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -28.41 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7888531 120 + 22407834 GCCCUUGAAGAAAGCUGCGUUGAUCAUGACCGCACUUUCAUCGGGCGUCAAGUCACUGGGUAUUAUAAUGUCUUUCACAUUAUCUAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCA ......((.(((((.((((((......)).))))))))).))(((((.(.((.(((((((((.....((((.....))))))))))))).)).).)((((........))))...)))). ( -33.70) >DroSec_CAF1 61161 120 + 1 GCCCUUGAAGAAAGCUGCGUUGAUCAUGACCCCACUUUCAUCGGGCGUCAAGUCACUGGGUGUUAGAAUGUAUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCA ((...((((((..(.(((.((((((.(((((((.........))).))))((.(((((((((...(((.....)))....))))))))).))))))))))).)...)).))))..))... ( -34.20) >DroSim_CAF1 62056 120 + 1 GCCCUUGAAGAAAGCUGCGUUGAUCAUGACCGCACUUUCAUCGGGCGUCAAGUCACUGGGUGUUAGAAUGUAUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCA ......((.(((((.((((((......)).))))))))).))(((((.(.((.(((((((((...(((.....)))....))))))))).)).).)((((........))))...)))). ( -36.90) >DroEre_CAF1 61246 120 + 1 GCCCUUGAAGAAAGCUGCGUUGAUCAUGACCGCACUUUCAUCGGGCGCCAGAUCACUGGGUGUCAUAAUAGCUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAAUCCA ((((..((.(((((.((((((......)).))))))))).))))))((((((.(((((((((..................)))))))))))))))(((........)))........... ( -39.17) >DroYak_CAF1 62568 120 + 1 GCCCUUGAAGAAAGCUGCGUUGAUCAUGACCGCACUUUCAUCGGGGGUCAAGUCACUGGGUGUUACAAUGUCUUUCACAUUAUCGAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCU ((((((((.(((((.((((((......)).))))))))).))))))))........(((((((...(((((.....))))).((((.(....).)))))))))))............... ( -37.40) >consensus GCCCUUGAAGAAAGCUGCGUUGAUCAUGACCGCACUUUCAUCGGGCGUCAAGUCACUGGGUGUUAGAAUGUCUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCA ((((..((.(((((.((((((......)).))))))))).))))))....((.(((((((((..................))))))))).))(((.((((........))))...))).. (-28.41 = -28.45 + 0.04)

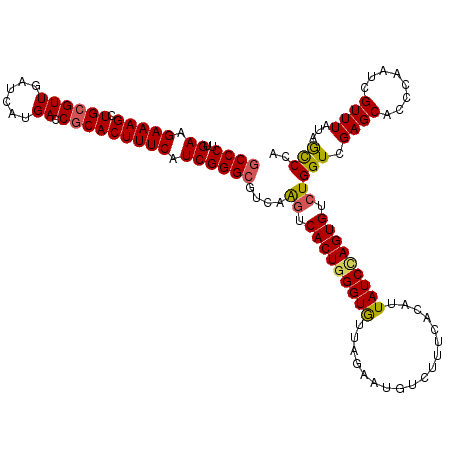
| Location | 7,888,571 – 7,888,691 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -37.61 |
| Consensus MFE | -30.35 |
| Energy contribution | -30.51 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7888571 120 + 22407834 UCGGGCGUCAAGUCACUGGGUAUUAUAAUGUCUUUCACAUUAUCUAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCAGGCUGCCUUCAAUCGAUCGGCCAGCUUAAUGGCUUCUGCC ..(((((.(.((.(((((((((.....((((.....))))))))))))).)).).)((((........))))...))))((((((((.((....))..)).))))))...(((....))) ( -35.70) >DroSec_CAF1 61201 120 + 1 UCGGGCGUCAAGUCACUGGGUGUUAGAAUGUAUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCAGGCUGCCUUCGAUCGAUCGGCCAGAUUAAUGGCCUCUGCC ..(((.((((...(((((((((...(((.....)))....)))))))))(((((((((.....(.((((....((((....))))....)))).).)))))))))....))))))).... ( -40.90) >DroSim_CAF1 62096 120 + 1 UCGGGCGUCAAGUCACUGGGUGUUAGAAUGUAUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCAGGCUGCCUUCGAUCUAUCGGCCAGAUUAAUGGCCUCUGCC ..(((.((((...(((((((((...(((.....)))....)))))))))(((((((((.......((((....((((....))))....))))...)))))))))....))))))).... ( -40.80) >DroEre_CAF1 61286 120 + 1 UCGGGCGCCAGAUCACUGGGUGUCAUAAUAGCUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAAUCCAGGCUGCCUUUGAUCGAUCGGCCAGCUUAAUAGCCUCUGCC ..((((((((((.(((((((((..................)))))))))))))))(((........)))..........((((((((...........)).))))))....))))..... ( -35.47) >DroYak_CAF1 62608 120 + 1 UCGGGGGUCAAGUCACUGGGUGUUACAAUGUCUUUCACAUUAUCGAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCUUGCUGCUUUUGAUCGAUCGGCCAACUUAAUGGCCUCUGCC .(((((((((((.((((.((((.....((((.....)))))))).)))).))((((((.....(.((((....((((....))))....)))).).)))))).......))))))))).. ( -35.20) >consensus UCGGGCGUCAAGUCACUGGGUGUUAGAAUGUCUUUCACAUUAUCCAGUGUCUGGUCGAGCACCCAAUCGUUUAUAGCCCAGGCUGCCUUCGAUCGAUCGGCCAGCUUAAUGGCCUCUGCC .((((.((((...(((((((((..................))))))))).((((((((.......((((....((((....))))....))))...)))))))).....)))).)))).. (-30.35 = -30.51 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:10 2006