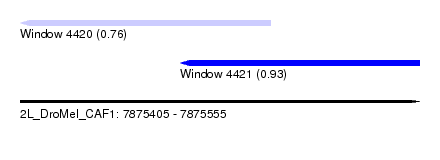
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,875,405 – 7,875,555 |
| Length | 150 |
| Max. P | 0.925820 |

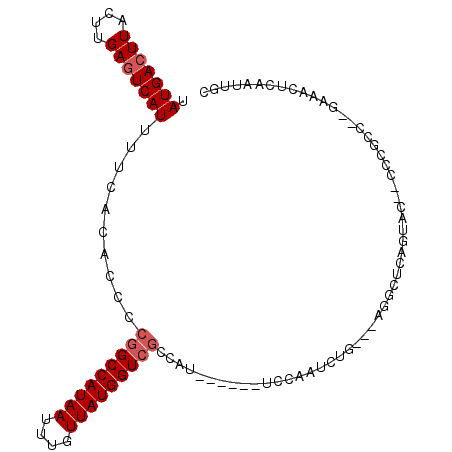
| Location | 7,875,405 – 7,875,499 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7875405 94 - 22407834 UAUGACUUACUUGAGUCAUUUUCACACCCCGGCCAUAAUUUGUUAUGGUCGCCAU------UCCAAUCUG---AGGCUCAGUAC--CCCGCC--GAAACUCAAUUGC .(((((((....)))))))..........(((((((((....)))))))))....------..((((.((---((..((.((..--...)).--))..)))))))). ( -24.40) >DroSec_CAF1 47233 83 - 1 UAUGACUUACUUGAGUCAUUUUCACACCCCGGCCAUAAUUUGUUAUGGUCGCCAU------UCCAAUCUG---AGGCUCAGUA---------------CUCAAUUGC .(((((((....)))))))..........(((((((((....)))))))))....------..((((.((---((........---------------)))))))). ( -22.00) >DroSim_CAF1 49000 83 - 1 UAUGACUUACUUGAGUCAUUUUCACACCCCGGCCAUAAUUUCUUAUGGUCGCCAU------UCCAAUUUG---AGGCUCGGUA---------------CUCAAUUGC .(((((((....)))))))..........(((((((((....)))))))))....------..(((((.(---((........---------------)))))))). ( -21.70) >DroEre_CAF1 48135 96 - 1 UAUGACUUACUUGACUCAUUUUCACACCCCUGCCAUAAUUUGUUAUGGUCGCCAU------UCCAAUCCG---AGGCUCAGUAAUUCCCGCC--GAAACUCAAUUGU ...((.(((((.((.................(((((((....))))))).(((..------.........---.)))))))))).)).....--............. ( -13.20) >DroYak_CAF1 49358 104 - 1 UAUGACUUACUUGAGUCAUUUUCACACCCCGGCCAUAAUUUGUUAUGGUCGCCAUUUCAAUUCCAAUCUG---AGGCUCAGUACCCCCCGUCCGGCAACUCAAUUGC .(((((((....)))))))..........(((((((((....)))))))))............((((.((---((.....(.((.....)).).....)))))))). ( -21.30) >DroAna_CAF1 49357 98 - 1 UAUGACUUACUCGAGUCAUUUUCACACCCCGGCCAUAAUUUGUUAUGGUCUCCGA------AUCAAUCUCGGUAAUCUCAGUAC--CCCGCC-GAAAACCCAAUUGC ..((((((....))))))(((((.......((((((((....)))))))).((((------.......))))............--......-)))))......... ( -20.60) >consensus UAUGACUUACUUGAGUCAUUUUCACACCCCGGCCAUAAUUUGUUAUGGUCGCCAU______UCCAAUCUG___AGGCUCAGUAC__CCCGCC__GAAACUCAAUUGC .(((((((....)))))))..........(((((((((....)))))))))........................................................ (-14.15 = -14.65 + 0.50)


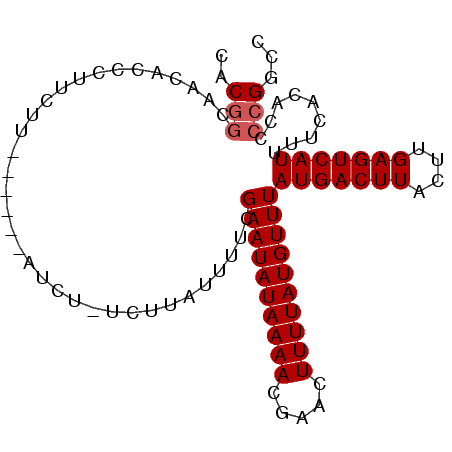
| Location | 7,875,465 – 7,875,555 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -16.78 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7875465 90 - 22407834 CACGGCUACACCCUUCAU------AUCU-UCUUAUUUUCGAAUAUAAAACGAACUUUUAUGUUUAUGACUUACUUGAGUCAUUUUCACACCCCGGCC ...((((...........------....-..........((((((((((.....))))))))))(((((((....)))))))...........)))) ( -15.70) >DroSec_CAF1 47282 90 - 1 CACGGCAACACCCUUCUU------AUCU-UCUUAUUUUCGAAUAUAAAACGAACUUUUAUGUUUAUGACUUACUUGAGUCAUUUUCACACCCCGGCC ...(((............------....-..........((((((((((.....))))))))))(((((((....)))))))............))) ( -14.80) >DroSim_CAF1 49049 90 - 1 CACGGCAACACCCUUCUU------AUCU-UCUUAUUUUCGAAUAUAAAACGAACUUUUAUGUUUAUGACUUACUUGAGUCAUUUUCACACCCCGGCC ...(((............------....-..........((((((((((.....))))))))))(((((((....)))))))............))) ( -14.80) >DroYak_CAF1 49428 90 - 1 CACGCCAACACCCUUCUU------AUCU-UCUUAUUUUCGAAUAUAAAACGAACUUUUAUGUUUAUGACUUACUUGAGUCAUUUUCACACCCCGGCC ...(((............------....-..........((((((((((.....))))))))))(((((((....)))))))...........))). ( -15.90) >DroAna_CAF1 49421 92 - 1 UACAGC-----CCUCCAUUCGGCCAUCGGUGCGUUUUUCGAAUAUAAAACGAACUUUUAUGUUUAUGACUUACUCGAGUCAUUUUCACACCCCGGCC ......-----.........((((...((((((((((........))))))........((...(((((((....)))))))...))))))..)))) ( -22.70) >consensus CACGGCAACACCCUUCUU______AUCU_UCUUAUUUUCGAAUAUAAAACGAACUUUUAUGUUUAUGACUUACUUGAGUCAUUUUCACACCCCGGCC ..(((..................................((((((((((.....))))))))))(((((((....))))))).........)))... (-11.82 = -12.22 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:05 2006