| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,852,654 – 7,852,745 |
| Length | 91 |
| Max. P | 0.629498 |

| Location | 7,852,654 – 7,852,745 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -6.22 |
| Energy contribution | -7.22 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.35 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7852654 91 + 22407834 UACUGUGUGUCAACAGU-------AACCAAUCAAGUAUUUAGUUUACUUGGUAACGGUUGCUCUUGUUU---------GUUAUUAUUUAAACAAAUUUGCUAUUUAC .(((((......)))))-------((((.((((((((.......))))))))...))))((....((((---------(((........)))))))..))....... ( -20.70) >DroSec_CAF1 24782 100 + 1 UACUUUGUGUCAACAGU-------AACCAAUCAAGUAUUAAGUUUACUUGGUAACGGUUGCUCUUGUUUGGCUUUGCAGUUAUUAUUUAACCAAAUUUCUUAUUUAC ...((((.(((((((((-------((((.((((((((.......))))))))...))))))...)).)))))......((((.....))))))))............ ( -19.50) >DroSim_CAF1 26723 104 + 1 UACUUUGUGUCAACAGUAAACAGUAAACAAUCAAGUAUUA-GUUUACUUGGUAACUGUUACUCUUGCUUGACUUAGCAGUUAUUAUUUAAACAAAUUUCCUAUUA-- (((((..(((..((........))..)))...)))))...-(((((..((((((((((((.((......))..))))))))))))..))))).............-- ( -21.10) >DroEre_CAF1 24951 88 + 1 UACUUUGUGUCAACAGU-------AACCAAUCAAAUAUUUAGUUUACUUGGUAACGGUUGCCCUUGUUUAACUU------------GCAACAAAAUGUCCUAUUUAC ...(((((..((((.((-------.(((((..((((.....))))..))))).)).))))....(((.......------------))))))))............. ( -12.30) >DroYak_CAF1 25430 88 + 1 UACUUUGUGUCAACAGU-------UACCAAUCAAAUAUUUAGUUUACUUGGUAACGGUUGCCGUUGUUUAACUU------------UUAACAAAAUUUCCUGUUUCC ........(.((((.((-------((((((..((((.....))))..)))))))).)))).).(((((......------------..))))).............. ( -16.40) >consensus UACUUUGUGUCAACAGU_______AACCAAUCAAGUAUUUAGUUUACUUGGUAACGGUUGCUCUUGUUUAACUU____GUUAUUAUUUAACCAAAUUUCCUAUUUAC .....(((....))).........((((.((((((((.......))))))))...))))................................................ ( -6.22 = -7.22 + 1.00)

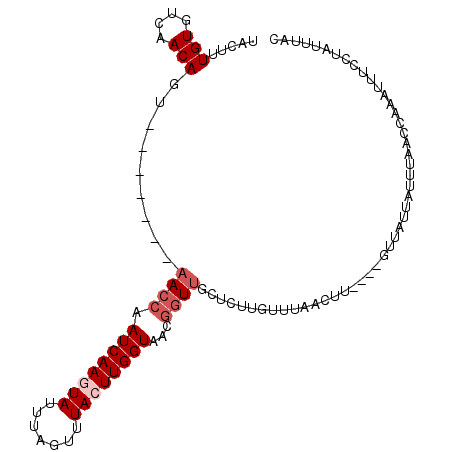

| Location | 7,852,654 – 7,852,745 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -17.93 |
| Consensus MFE | -6.10 |
| Energy contribution | -6.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
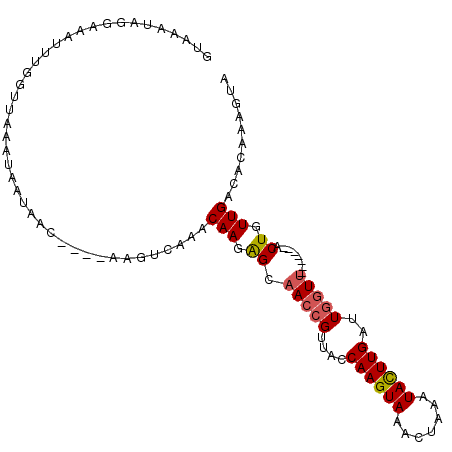
>2L_DroMel_CAF1 7852654 91 - 22407834 GUAAAUAGCAAAUUUGUUUAAAUAAUAAC---------AAACAAGAGCAACCGUUACCAAGUAAACUAAAUACUUGAUUGGUU-------ACUGUUGACACACAGUA .......((...((((((........)))---------)))(((.((.(((((....((((((.......))))))..)))))-------.)).))).......)). ( -16.60) >DroSec_CAF1 24782 100 - 1 GUAAAUAAGAAAUUUGGUUAAAUAAUAACUGCAAAGCCAAACAAGAGCAACCGUUACCAAGUAAACUUAAUACUUGAUUGGUU-------ACUGUUGACACAAAGUA ............(((((((...............)))))))(((.((.(((((....((((((.......))))))..)))))-------.)).))).......... ( -17.96) >DroSim_CAF1 26723 104 - 1 --UAAUAGGAAAUUUGUUUAAAUAAUAACUGCUAAGUCAAGCAAGAGUAACAGUUACCAAGUAAAC-UAAUACUUGAUUGUUUACUGUUUACUGUUGACACAAAGUA --..........(((((............((((......))))...((((((((.....(((((((-............)))))))....)))))).)))))))... ( -18.60) >DroEre_CAF1 24951 88 - 1 GUAAAUAGGACAUUUUGUUGC------------AAGUUAAACAAGGGCAACCGUUACCAAGUAAACUAAAUAUUUGAUUGGUU-------ACUGUUGACACAAAGUA ((.((((((((...(((((..------------......)))))((....)))))(((((.((((.......)))).))))).-------.))))).))........ ( -16.80) >DroYak_CAF1 25430 88 - 1 GGAAACAGGAAAUUUUGUUAA------------AAGUUAAACAACGGCAACCGUUACCAAGUAAACUAAAUAUUUGAUUGGUA-------ACUGUUGACACAAAGUA (....).....(((((((...------------..(((....)))(.((((.((((((((.((((.......)))).))))))-------)).)))).)))))))). ( -19.70) >consensus GUAAAUAGGAAAUUUGGUUAAAUAAUAAC____AAGUCAAACAAGAGCAACCGUUACCAAGUAAACUAAAUACUUGAUUGGUU_______ACUGUUGACACAAAGUA .........................................(((.((.(((((....((((((.......))))))..)))))........)).))).......... ( -6.10 = -6.22 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:56 2006