
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,851,532 – 7,851,665 |
| Length | 133 |
| Max. P | 0.993683 |

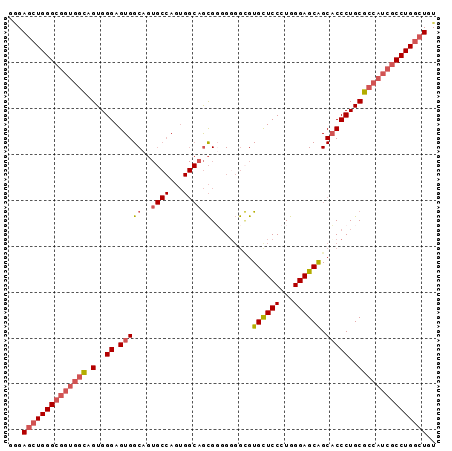
| Location | 7,851,532 – 7,851,625 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -48.02 |
| Consensus MFE | -34.22 |
| Energy contribution | -36.35 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7851532 93 - 22407834 GAGAACUGGGCGGUGGUAGUGGGAGUGGCAGUGCCAGUGGCAGCGGUGUGGCGUGUUCCCUGGGAGCAGCACCCUGCGCCAUCGCCUGGCUGU .....(..(((((((((......(.((((...)))).).((((.(((((....((((((...))))))))))))))))))))))))..).... ( -44.50) >DroSec_CAF1 23683 93 - 1 GGGAGCUGGGCGGUGUCAGUGGGAGUGGCAGUGCCAGUGGCAGCGUGGGGGGGUGCUCCUUGGGAGCAGCACCCUGCGCCAUCGCCUGGCUGU ...(((..((((((((((.(((.(.......).))).)))).(((..(((.(.((((((...)))))).).)))..))).))))))..))).. ( -53.10) >DroSim_CAF1 25614 93 - 1 GGGAGCUGGGCGGUGGCAGUGGGAGUGGCAGUGCCAGUGGCAGCGUGGGGGCGUGUUCCCUGGGAGCAGCACCCUGCGCCAUCGCCUGGCUGU ...(((..(((((((((..((..(.((((...)))).)..))..(..(((((.((((((...)))))))).)))..))))))))))..))).. ( -52.90) >DroAna_CAF1 25705 81 - 1 GCGAGGUGGGCGCCAGCAGUGGGAGUGGC------------CAUGGGGGUAUGUGCUCCCUGGGAGCGGCCCCCUGCGCCAUAUCCUGGCUGU (((.......)))((((...(((.(((((------------((.((((((...((((((...)))))))))))))).))))).)))..)))). ( -41.60) >consensus GGGAGCUGGGCGGUGGCAGUGGGAGUGGCAGUGCCAGUGGCAGCGGGGGGGCGUGCUCCCUGGGAGCAGCACCCUGCGCCAUCGCCUGGCUGU ...((((((((((((((.(..((.(((((..((((...)))))).........((((((...)))))).)))))..))))))))))))))).. (-34.22 = -36.35 + 2.13)


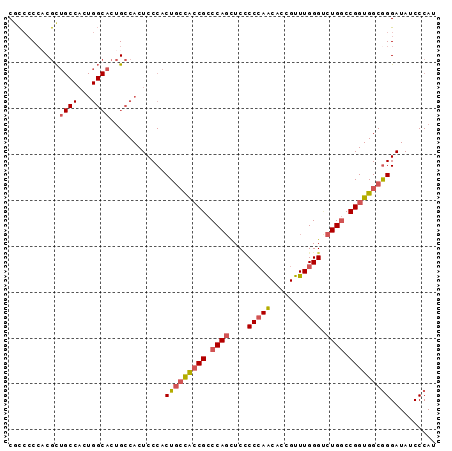
| Location | 7,851,572 – 7,851,665 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -21.86 |
| Energy contribution | -23.05 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7851572 93 + 22407834 CGCCACACCGCUGCCACUGGCACUGCCACUCCCACUACCACCGCCCAGUUCUCCCAACACCGUUUGGGCCUGGCCGGUGGCGGGAUAUCCCAU .((((............))))................((((((.((((....(((((......))))).)))).)))))).(((....))).. ( -31.80) >DroSec_CAF1 23723 93 + 1 CCCCCCCACGCUGCCACUGGCACUGCCACUCCCACUGACACCGCCCAGCUCCCCCAACACCGUUUGGGUCUGGCCGGUGGCGGGAUAUCCCAU ............(((((((((.............(((........)))....(((((......)))))....)))))))))(((....))).. ( -31.40) >DroSim_CAF1 25654 93 + 1 CGCCCCCACGCUGCCACUGGCACUGCCACUCCCACUGCCACCGCCCAGCUCCCCCAACACCGUUUGGGUCUGGCCGGUGGCGGGAUAUCCCAU ....(((..((((((...))))..))..........(((((((.((((....(((((......))))).)))).))))))))))......... ( -36.30) >DroAna_CAF1 25745 81 + 1 CAUACCCCCAUG------------GCCACUCCCACUGCUGGCGCCCACCUCGCCGAGCACCGUUUGGGACUGCCCGGUGGCAGGAUAUCCCAU ....((.(((((------------(.((.(((((.((((((((.......)))).)))).....))))).)).)).))))..))......... ( -25.70) >consensus CGCCCCCACGCUGCCACUGGCACUGCCACUCCCACUGCCACCGCCCAGCUCCCCCAACACCGUUUGGGUCUGGCCGGUGGCGGGAUAUCCCAU ...........((((...))))............(((((((((.((((....(((((......))))).)))).))))))))).......... (-21.86 = -23.05 + 1.19)


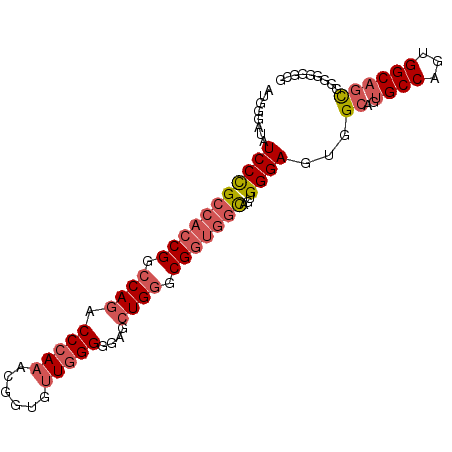
| Location | 7,851,572 – 7,851,665 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -25.55 |
| Energy contribution | -27.68 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7851572 93 - 22407834 AUGGGAUAUCCCGCCACCGGCCAGGCCCAAACGGUGUUGGGAGAACUGGGCGGUGGUAGUGGGAGUGGCAGUGCCAGUGGCAGCGGUGUGGCG ........(((((((((((.((((.(((((......)))))....)))).)))))))...))))((.(((.(((........))).))).)). ( -44.70) >DroSec_CAF1 23723 93 - 1 AUGGGAUAUCCCGCCACCGGCCAGACCCAAACGGUGUUGGGGGAGCUGGGCGGUGUCAGUGGGAGUGGCAGUGCCAGUGGCAGCGUGGGGGGG .........(((.((.((((((...(((((......))))).).)))))(((.(((((.(((.(.......).))).))))).))))).))). ( -41.40) >DroSim_CAF1 25654 93 - 1 AUGGGAUAUCCCGCCACCGGCCAGACCCAAACGGUGUUGGGGGAGCUGGGCGGUGGCAGUGGGAGUGGCAGUGCCAGUGGCAGCGUGGGGGCG ..(((....)))(((((((.((((.(((((......)))))....)))).)))))))..((..(.((((...)))).)..))((......)). ( -43.00) >DroAna_CAF1 25745 81 - 1 AUGGGAUAUCCUGCCACCGGGCAGUCCCAAACGGUGCUCGGCGAGGUGGGCGCCAGCAGUGGGAGUGGC------------CAUGGGGGUAUG .....((((((..(((..((.((.(((((...((((((((......)))))))).....))))).)).)------------).))))))))). ( -35.90) >consensus AUGGGAUAUCCCGCCACCGGCCAGACCCAAACGGUGUUGGGGGAGCUGGGCGGUGGCAGUGGGAGUGGCAGUGCCAGUGGCAGCGGGGGGGCG ........(((((((((((.((((.(((((......)))))....)))).)))))))...))))...((..((((...))))))......... (-25.55 = -27.68 + 2.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:54 2006