
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
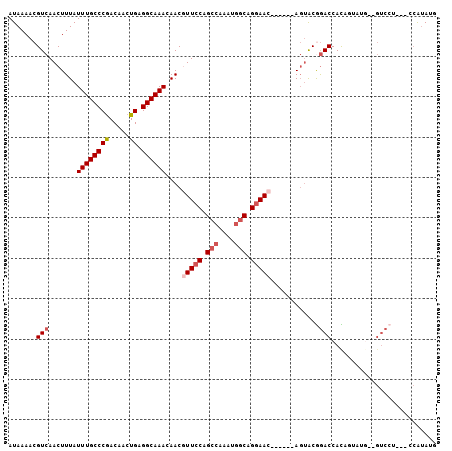
| Location | 7,844,718 – 7,844,813 |
| Length | 95 |
| Max. P | 0.869693 |

| Location | 7,844,718 – 7,844,813 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -15.77 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
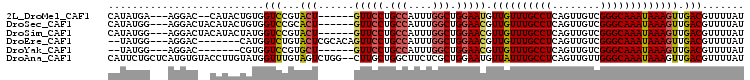
>2L_DroMel_CAF1 7844718 95 + 22407834 AUAAAACGUCAACUUUAUUUGCCCGACAACUGAGGCAAACAACAUUCCAGCCAAAUGGCAGGAAC------AGUACGGACCACAGUAUG--GUCCU---UCAUAUG ...........(((...((((((((.....)).)))))).....((((.(((....))).)))).------)))..((((((.....))--)))).---....... ( -24.80) >DroSec_CAF1 16830 97 + 1 AUAAAACGUCAACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAAC------AGUGCGGACCACAGUAUGUAGUCCU---CCAUAUG .................((((((((.....)).))))))....(((((.(((....))).)))))------.(((.((((.((.....)).)))).---.)))... ( -26.60) >DroSim_CAF1 17995 97 + 1 AUAAAACGUCAACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAAC------AGUACGGACCAUAGUAUGUAGUCCU---CCAUAUG .......(((.(((...((((((((.....)).))))))....(((((.(((....))).)))))------)))...)))....(((((.......---.))))). ( -24.10) >DroEre_CAF1 17074 94 + 1 AUAAAACGUCAACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAACUGUGCGAGUACAGACCAUG-------GUCCU---CCAUA-- .......(((.......((((((((.....)).))))))....(((((.(((....))).)))))((((....))))))).(((-------(....---)))).-- ( -25.90) >DroYak_CAF1 17366 88 + 1 AUAAAACGUCAACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAAC------AGCACGGACCACG-------GUCCU---CCAUA-- .......(((.......((((((((.....)).))))))....(((((.(((....))).)))))------.).))((((....-------)))).---.....-- ( -26.10) >DroAna_CAF1 18689 104 + 1 AUAAAACGUCAACUUUAUUUGCCCAACAACUGAGGCAAAUAACAUUCCAGCGAGAAGCCAGCAAG--CCAGACUACAAACCAUACAAGGUACACAUGAGCAGAAUG ..............(((((((((((.....)).)))))))))(((((..((.........))..(--(((........(((......))).....)).)).))))) ( -19.02) >consensus AUAAAACGUCAACUUUAUUUGCCCGACAACUGAGGCAAACAACGUUCCAGCCAAAUGGCAGGAAC______AGUACGGACCACAGUAUG__GUCCU___CCAUAUG .......(((.......((((((((.....)).))))))....(((((.(((....))).)))))............))).......................... (-15.77 = -16.63 + 0.86)


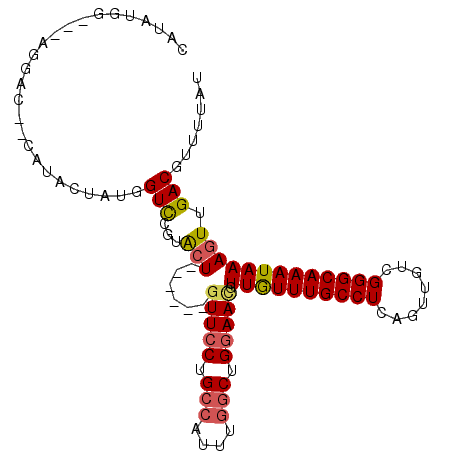
| Location | 7,844,718 – 7,844,813 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
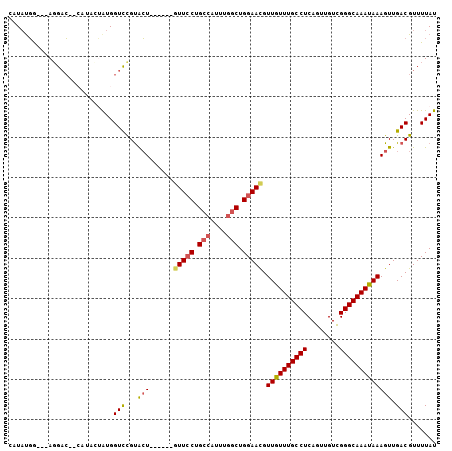
>2L_DroMel_CAF1 7844718 95 - 22407834 CAUAUGA---AGGAC--CAUACUGUGGUCCGUACU------GUUCCUGCCAUUUGGCUGGAAUGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAU .......---.((((--(((...)))))))(((((------(((((.(((....))).))))).((((((((((........)))))))))))))..))....... ( -32.90) >DroSec_CAF1 16830 97 - 1 CAUAUGG---AGGACUACAUACUGUGGUCCGCACU------GUUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAU .......---.(((((((.....)))))))..(((------(((((.(((....))).))))).((((((((((........)))))))))))))........... ( -35.40) >DroSim_CAF1 17995 97 - 1 CAUAUGG---AGGACUACAUACUAUGGUCCGUACU------GUUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAU .......---.((((((.......))))))(((((------(((((.(((....))).))))).((((((((((........)))))))))))))..))....... ( -31.70) >DroEre_CAF1 17074 94 - 1 --UAUGG---AGGAC-------CAUGGUCUGUACUCGCACAGUUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAU --(((((---....)-------))))(((...(((......(((((.(((....))).))))).((((((((((........))))))))))))).)))....... ( -31.40) >DroYak_CAF1 17366 88 - 1 --UAUGG---AGGAC-------CGUGGUCCGUGCU------GUUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAU --.....---.((((-------....))))(((((------(((((.(((....))).))))).((((((((((........)))))))))))))..))....... ( -32.00) >DroAna_CAF1 18689 104 - 1 CAUUCUGCUCAUGUGUACCUUGUAUGGUUUGUAGUCUGG--CUUGCUGGCUUCUCGCUGGAAUGUUAUUUGCCUCAGUUGUUGGGCAAAUAAAGUUGACGUUUUAU ((((((((........(((......))).....(...((--((....))))...))).))))))((((((((((........)))))))))).............. ( -23.30) >consensus CAUAUGG___AGGAC__CAUACUAUGGUCCGUACU______GUUCCUGCCAUUUGGCUGGAACGUUGUUUGCCUCAGUUGUCGGGCAAAUAAAGUUGACGUUUUAU ..........................(((...(((......(((((.(((....))).))))).((((((((((........))))))))))))).)))....... (-22.51 = -22.82 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:51 2006