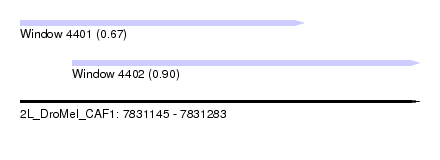
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,831,145 – 7,831,283 |
| Length | 138 |
| Max. P | 0.896520 |

| Location | 7,831,145 – 7,831,243 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.88 |
| Mean single sequence MFE | -17.16 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7831145 98 + 22407834 AAGACCCGAUUAAAACUAAGCUGAACAAAAAUUGUCUGCUCAAAAGCAUGCAAAUAUAUUCUAGAUAAAUUUUUUAUUGAUCUUAAUAGGAAAUUUUA ...............................((((.((((....)))).)))).............(((((((((((((....))))))))))))).. ( -12.50) >DroSec_CAF1 3108 98 + 1 AAGACUCUAUUAAAACUAAGCUGGUCAAAAAUUGUGUGCUCAAAAGCAUGCAAAUAUAUUCCAGAUAUAUUUUUUAUUGAUGUUAAUAGGAAAUUUUA .....((((((((.......((((.......(((((((((....))))))))).......))))...((((.......))))))))))))........ ( -18.84) >DroSim_CAF1 3250 98 + 1 AAAACUCGAUUAAAACUAAGCUGGUCAAAAAUUGUGUGCUCAAAAGCAUGCAAAUAUAUUCCAGAUAUAUUUUUUAUUGAUGUUAAUAGGAAAUUUUA ..((((((((.((((.((..((((.......(((((((((....))))))))).......))))...)).)))).))))).))).............. ( -20.14) >consensus AAGACUCGAUUAAAACUAAGCUGGUCAAAAAUUGUGUGCUCAAAAGCAUGCAAAUAUAUUCCAGAUAUAUUUUUUAUUGAUGUUAAUAGGAAAUUUUA ....................((((.......(((((((((....))))))))).......))))....(((((((((((....))))))))))).... (-16.43 = -16.87 + 0.45)


| Location | 7,831,163 – 7,831,283 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -16.26 |
| Energy contribution | -19.08 |
| Covariance contribution | 2.82 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7831163 120 + 22407834 AGCUGAACAAAAAUUGUCUGCUCAAAAGCAUGCAAAUAUAUUCUAGAUAAAUUUUUUAUUGAUCUUAAUAGGAAAUUUUAUCGGGGCAUGCACUUGAUGAAAAUCAAUAAUCAGUAAUCG .(((((.....................((((((............((((((((((((((((....)))))))))).))))))...))))))..(((((....)))))...)))))..... ( -26.96) >DroSec_CAF1 3126 120 + 1 AGCUGGUCAAAAAUUGUGUGCUCAAAAGCAUGCAAAUAUAUUCCAGAUAUAUUUUUUAUUGAUGUUAAUAGGAAAUUUUAUCGGGGCUUCCACUUGAUAAAAAUCAAUAAUCAGUAAUCG ..((((.......(((((((((....))))))))).......))))...((((..((((((((............((((((((((.......))))))))))))))))))..)))).... ( -28.64) >DroSim_CAF1 3268 120 + 1 AGCUGGUCAAAAAUUGUGUGCUCAAAAGCAUGCAAAUAUAUUCCAGAUAUAUUUUUUAUUGAUGUUAAUAGGAAAUUUUAUCGGGGCUUGCACUUGAUGAAAAUCAAUAAUCAGUAAUCG .((.((((.....(((((((((....))))))))).......((.((((.(((((((((((....)))))))))))..)))))))))).))..(((((....)))))............. ( -30.50) >DroEre_CAF1 3308 118 + 1 UACUGAAACAAAA--GUGUGCUGAAAAUCGUGAAAAUAUAUUCAAGAUAUAUUUUUUAUUACUCCUAAUAGGAAAUUUUAUCUGGGCAUGCACUCGAUGAAAAUCAAUAAUCAGUAAUCG ((((((.......--(((((((........((((......))))(((((.(((((((((((....)))))))))))..))))).)))))))....(((....))).....)))))).... ( -28.30) >DroYak_CAF1 3264 105 + 1 ---------------UUGUGCUCUACAUCAUGCAAAUGUUUUCUAGAUAUAUUUUUUAACGAUUUUAAUAGGAAAUUUUAUCUGGGCAUGCACUUGGUGGAAAUCAAUAAUGAGUAAUCG ---------------...(((((.......((((..(((...(((((((.((((((((..........))))))))..)))))))))))))).(((((....)))))....))))).... ( -21.20) >consensus AGCUGAACAAAAAUUGUGUGCUCAAAAGCAUGCAAAUAUAUUCCAGAUAUAUUUUUUAUUGAUCUUAAUAGGAAAUUUUAUCGGGGCAUGCACUUGAUGAAAAUCAAUAAUCAGUAAUCG .(((((.......(((((((((....)))))))))..........(((...((((((((((....))))))))))((((((((((.......))))))))))))).....)))))..... (-16.26 = -19.08 + 2.82)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:47 2006