
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 799,872 – 800,006 |
| Length | 134 |
| Max. P | 0.982242 |

| Location | 799,872 – 799,977 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.70 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 799872 105 + 22407834 ACAAAAGAUUUCAUUAAUACUGUUUUUA-----------UUCCUAGGUAACAGAUGCUGUGA----CCCAACUGCCGGGAGAAAGUGCACCUGGGCCCAUUAACAUUUUAAUGGGUUAUU ............................-----------..(((((((.((((...))))..----(((.......))).........)))))))((((((((....))))))))..... ( -23.70) >DroSec_CAF1 59600 105 + 1 AAAUAAGAUUUCAUUAAUACUGUUUCUA-----------UUCCUAGGUCACAGAUGCUGUGA----CCCAACUGCCGGGCGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUU ............................-----------......((((((((...))))))----))......(((((.(......).)))))(((((((((....))))))))).... ( -31.80) >DroSim_CAF1 64199 105 + 1 AAAUAAGAUUUCAUUAAUACUGUUCUUA-----------UUCCUAGGUCACAGAUGCUGUGA----CCCAAGUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACGUUUUAAUGGGUCAUU .(((((((...((.......)).)))))-----------))....((((((((...))))))----))......(((((.(......).)))))(((((((((....))))))))).... ( -34.10) >DroYak_CAF1 63720 107 + 1 ---UAAGAUUUUAUUGAUCCUGUUUUUAAUAGCUCCUUGCACCUUGGUCACAGAUGCUGGGCGGAGUGCAA------GGUG----GUCACCUGGGCCGCUUAAGAUUUUAAUGGGUUAUU ---...........((((((.(((((....(((.((((((((.((.(((.(((...)))))).))))))))------)).(----(((.....))))))).)))))......)))))).. ( -32.30) >consensus AAAUAAGAUUUCAUUAAUACUGUUUUUA___________UUCCUAGGUCACAGAUGCUGUGA____CCCAACUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUU ................................................(((((...)))))..............((((.(......).))))((((((((((....))))))))))... (-14.57 = -15.70 + 1.13)


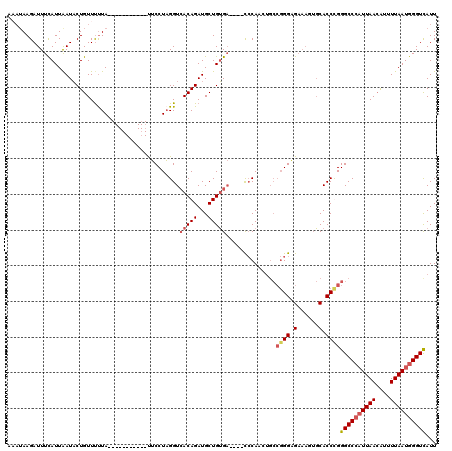
| Location | 799,872 – 799,977 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -13.71 |
| Energy contribution | -17.15 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 799872 105 - 22407834 AAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCAGUUGGG----UCACAGCAUCUGUUACCUAGGAA-----------UAAAAACAGUAUUAAUGAAAUCUUUUGU .......((((((.(((((....(((((((.(((.(........)))).)))))----))..)))))(((((.........-----------....)))))..))))))........... ( -23.52) >DroSec_CAF1 59600 105 - 1 AAUGACCCAUUAAAAUGUUAAUGGGCCCGGGUGCACUUUCGCCCGGCAGUUGGG----UCACAGCAUCUGUGACCUAGGAA-----------UAGAAACAGUAUUAAUGAAAUCUUAUUU .....((((((((....)))))))).(((((((......)))))))...(((((----((((((...)))))))))))...-----------............................ ( -35.40) >DroSim_CAF1 64199 105 - 1 AAUGACCCAUUAAAACGUUAAUGGGCCCGGGUGCACUUUCUCCCGGCACUUGGG----UCACAGCAUCUGUGACCUAGGAA-----------UAAGAACAGUAUUAAUGAAAUCUUAUUU .....((((((((....)))))))).(((((.(......).)))))..((((((----((((((...))))))))))))((-----------(((((.((.......))...))))))). ( -37.60) >DroYak_CAF1 63720 107 - 1 AAUAACCCAUUAAAAUCUUAAGCGGCCCAGGUGAC----CACC------UUGCACUCCGCCCAGCAUCUGUGACCAAGGUGCAAGGAGCUAUUAAAAACAGGAUCAAUAAAAUCUUA--- ((((.(((.............((((.(.((((...----.)))------).)....))))...((((((.......))))))..)).).))))......(((((.......))))).--- ( -20.30) >consensus AAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCAGUUGGG____UCACAGCAUCUGUGACCUAGGAA___________UAAAAACAGUAUUAAUGAAAUCUUAUUU .....((((((((....)))))))).(((((((......)))))))...(((((....((((((...))))))))))).......................................... (-13.71 = -17.15 + 3.44)


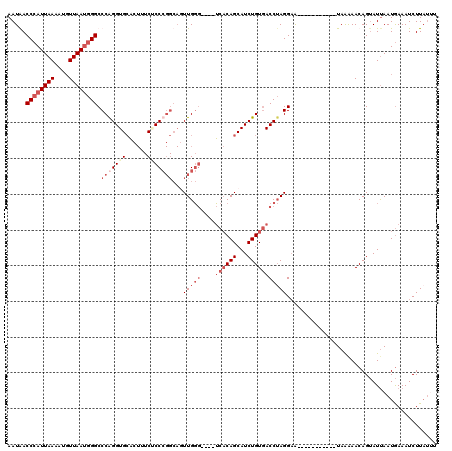
| Location | 799,901 – 800,006 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -19.70 |
| Energy contribution | -21.62 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 799901 105 + 22407834 UCCUAGGUAACAGAUGCUGUGA----CCCAACUGCCGGGAGAAAGUGCACCUGGGCCCAUUAACAUUUUAAUGGGUUAUUAUUUGUCUCUGUGCAUCUGCAUCUGCUGG ...(((((..(((((((.(.((----(.......(((((.(......).)))))(((((((((....)))))))))........))).)...))))))).))))).... ( -33.10) >DroSec_CAF1 59629 105 + 1 UCCUAGGUCACAGAUGCUGUGA----CCCAACUGCCGGGCGAAAGUGCACCCGGGCCCAUUAACAUUUUAAUGGGUCAUUAUUUGUCUCUGUGCAUCUGCAUCUGUCGG ...(((((..(((((((.(.((----(........((((.(......).))))((((((((((....)))))))))).......))).)...))))))).))))).... ( -36.30) >DroSim_CAF1 64228 105 + 1 UCCUAGGUCACAGAUGCUGUGA----CCCAAGUGCCGGGAGAAAGUGCACCCGGGCCCAUUAACGUUUUAAUGGGUCAUUAUUUGUCUCUGUGCAUCUGCAUCUGUCGG ...(((((..(((((((...((----..((((((.((((.(......).))))((((((((((....))))))))))..))))))..))...))))))).))))).... ( -38.50) >DroEre_CAF1 60874 87 + 1 UCCGUGGCCACAGAUGCUGUGA----CCCAA----------------CUCCUGGG--GAUUAAGACUUUGAUGGCUUAUUAUUUGUCUCUGUGCAUCUGCAUCUGGAGG .....(((((((((..((....----((((.----------------....))))--.....))..)))).)))))..........((((((((....))))..)))). ( -23.30) >DroYak_CAF1 63757 99 + 1 ACCUUGGUCACAGAUGCUGGGCGGAGUGCAA------GGUG----GUCACCUGGGCCGCUUAAGAUUUUAAUGGGUUAUUAUUUGUCUCCGUGCAUCUGCAUCUGGUGG ........(((((((((((.((((((.((((------((((----(((.....))))))))..(((((....)))))......))))))))).))...)))))).))). ( -35.00) >consensus UCCUAGGUCACAGAUGCUGUGA____CCCAA_UGCCGGGAGAAAGUGCACCUGGGCCCAUUAACAUUUUAAUGGGUUAUUAUUUGUCUCUGUGCAUCUGCAUCUGGCGG .((..((...(((((((..................((((.(......).))))((((((((((....))))))))))...............)))))))...))...)) (-19.70 = -21.62 + 1.92)

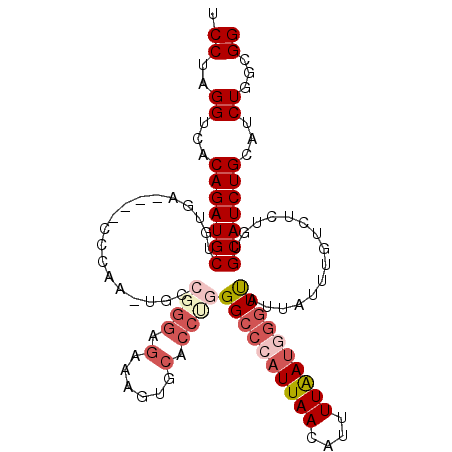

| Location | 799,901 – 800,006 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -20.67 |
| Energy contribution | -23.08 |
| Covariance contribution | 2.41 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 799901 105 - 22407834 CCAGCAGAUGCAGAUGCACAGAGACAAAUAAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCAGUUGGG----UCACAGCAUCUGUUACCUAGGA ((((.....((((((((.................((((((((....))))))))(((((.(((.(........)))).)))))----.....))))))))...)).)). ( -34.30) >DroSec_CAF1 59629 105 - 1 CCGACAGAUGCAGAUGCACAGAGACAAAUAAUGACCCAUUAAAAUGUUAAUGGGCCCGGGUGCACUUUCGCCCGGCAGUUGGG----UCACAGCAUCUGUGACCUAGGA ((......(((....)))....(((.........((((((((....)))))))).(((((((......)))))))..)))(((----((((((...))))))))).)). ( -38.30) >DroSim_CAF1 64228 105 - 1 CCGACAGAUGCAGAUGCACAGAGACAAAUAAUGACCCAUUAAAACGUUAAUGGGCCCGGGUGCACUUUCUCCCGGCACUUGGG----UCACAGCAUCUGUGACCUAGGA (((..(((...((.(((((.....((.....)).((((((((....)))))))).....))))))).)))..)))..((((((----((((((...)))))))))))). ( -37.80) >DroEre_CAF1 60874 87 - 1 CCUCCAGAUGCAGAUGCACAGAGACAAAUAAUAAGCCAUCAAAGUCUUAAUC--CCCAGGAG----------------UUGGG----UCACAGCAUCUGUGGCCACGGA .((((...(((....)))..(((((..................)))))....--....))))----------------(((((----((((((...)))))))).))). ( -22.87) >DroYak_CAF1 63757 99 - 1 CCACCAGAUGCAGAUGCACGGAGACAAAUAAUAACCCAUUAAAAUCUUAAGCGGCCCAGGUGAC----CACC------UUGCACUCCGCCCAGCAUCUGUGACCAAGGU ..(((.(.(((((((((..(....).........................((((.(.((((...----.)))------).)....))))...))))))))).)...))) ( -24.90) >consensus CCAACAGAUGCAGAUGCACAGAGACAAAUAAUAACCCAUUAAAAUGUUAAUGGGCCCAGGUGCACUUUCUCCCGGCA_UUGGG____UCACAGCAUCUGUGACCUAGGA ((.......((((((((.................((((((((....))))))))(((((.(((...........))).))))).........))))))))......)). (-20.67 = -23.08 + 2.41)

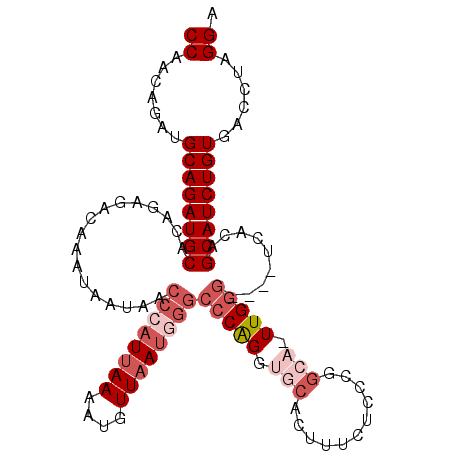

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:53 2006