| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,790,660 – 7,790,753 |
| Length | 93 |
| Max. P | 0.999728 |

| Location | 7,790,660 – 7,790,753 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.76 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -15.67 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7790660 93 + 22407834 UCAUGCUCAUUUGGCGGUUUUUUGGAAUUGAUCCGGGUUUCCCCAGUCCC------CUCCGCCGGAUGUUCUCUUCAC---------------GAACUAUGAAUUUUCCAUGUU---- (((((...(((((((((......(((.....)))(((....)))......------..)))))))))((((.......---------------)))))))))............---- ( -24.00) >DroSim_CAF1 3549 115 + 1 UCAUGCUCAUUUGGCGGUUUUU-CGAAUUGAUCCGGGUUUUCCCAGUCCCCGUCCACUCCGCCGGAUGUUCUGUUCACCAACUAUAUUUUGGAGGGCUAUGACUUUUCCUGCUUUG-- (((((((((((((((((.....-((....(((..(((....))).)))..))......)))))))))..........((((.......)))).)))).))))..............-- ( -29.10) >DroYak_CAF1 3628 111 + 1 UGAUGCACACUUGGCGGUUAUU-CGGUCUGAUCCGGGUUUUACCAGGCCCC-----CACCGCCGGAUGUUCUAAGCAUCAACUACUUUUUGGUGAGCUUGGGUUUUUCAUUA-AUAUC (((((.(((..(((((((....-.((......))(((((......))))).-----.)))))))..)))(((((((.(((.(((.....))))))))))))).....)))))-..... ( -33.60) >consensus UCAUGCUCAUUUGGCGGUUUUU_CGAAUUGAUCCGGGUUUUCCCAGUCCCC_____CUCCGCCGGAUGUUCUAUUCACCAACUA__UUUUGG_GAGCUAUGAAUUUUCCUUAUUU___ .......((((((((((.......(........)(((..........)))........)))))))))).................................................. (-15.67 = -16.00 + 0.33)


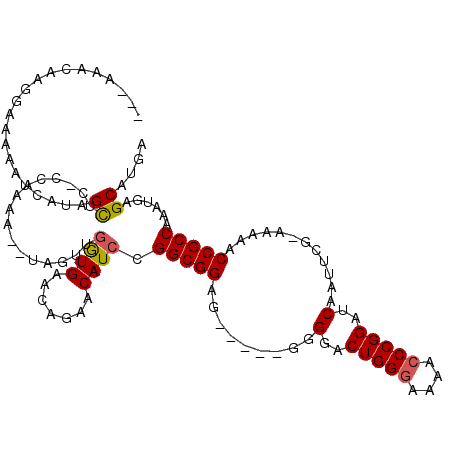
| Location | 7,790,660 – 7,790,753 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 66.76 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7790660 93 - 22407834 ----AACAUGGAAAAUUCAUAGUUC---------------GUGAAGAGAACAUCCGGCGGAG------GGGACUGGGGAAACCCGGAUCAAUUCCAAAAAACCGCCAAAUGAGCAUGA ----..((((.....(((((.((((---------------.......))))....(((((((------....))(((....)))(((.....)))......)))))..))))))))). ( -33.00) >DroSim_CAF1 3549 115 - 1 --CAAAGCAGGAAAAGUCAUAGCCCUCCAAAAUAUAGUUGGUGAACAGAACAUCCGGCGGAGUGGACGGGGACUGGGAAAACCCGGAUCAAUUCG-AAAAACCGCCAAAUGAGCAUGA --....((.(((....((........((((.......))))......))...)))(((((..((((..((..(((((....))))).))..))))-.....)))))......)).... ( -26.94) >DroYak_CAF1 3628 111 - 1 GAUAU-UAAUGAAAAACCCAAGCUCACCAAAAAGUAGUUGAUGCUUAGAACAUCCGGCGGUG-----GGGGCCUGGUAAAACCCGGAUCAGACCG-AAUAACCGCCAAGUGUGCAUCA .....-..............((((.((......))))))(((((.....((((..(((((((-----(.(..((((......))))..)...)).-....))))))..))))))))). ( -27.90) >consensus ___AAACAAGGAAAAAUCAUAGCUC_CCAAAA__UAGUUGGUGAACAGAACAUCCGGCGGAG_____GGGGACUGGGAAAACCCGGAUCAAUUCG_AAAAACCGCCAAAUGAGCAUGA .....................((................((((.......)))).(((((.........(..(((((....)))))..)............)))))......)).... (-18.14 = -18.37 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:38 2006