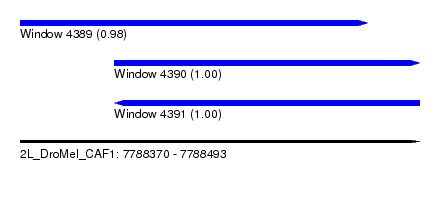
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,788,370 – 7,788,493 |
| Length | 123 |
| Max. P | 0.998021 |

| Location | 7,788,370 – 7,788,477 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -13.80 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
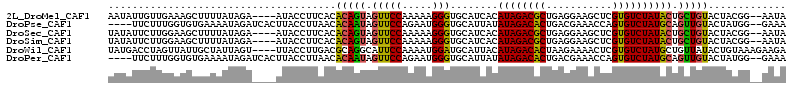
>2L_DroMel_CAF1 7788370 107 + 22407834 AAUAUUGUUGAAAGCUUUUAUAGA----AUACCUUCACACAGUAGUUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUA ..((((((((((.(..(((...))----)..).)))).))))))(((((......((((((.((.((((((((.(((....)))))))))))..)).))))))..))--))). ( -30.00) >DroPse_CAF1 1287 107 + 1 ----UUCUUUGGUGUGAAAAUAGAUCACUUACCUUAACACAAUAGUUCCAGAAUGGGUGCAUUAUAUAGACACUGACGAAACCAGUGUCUAUGCAGUUGUACUAUGG--GAAA ----......(((((((.......))))..)))............(((((.....((((((...(((((((((((.......)))))))))))....)))))).)))--)).. ( -30.70) >DroSec_CAF1 1273 107 + 1 UAUAUUCUUGGAAGCUUUUAUAGA----AUACCUUCACACAGUAGUUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUA ..(((((((((...........((----(....)))..(((((((((((.....)))........((((((((.(((....))))))))))))))))))).))).))--)))) ( -29.20) >DroSim_CAF1 1268 107 + 1 UAUAUUCUUGGAAGCUUUUAUAGA----AUACCUUCACACAGUAGUUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUA ..(((((((((...........((----(....)))..(((((((((((.....)))........((((((((.(((....))))))))))))))))))).))).))--)))) ( -29.20) >DroWil_CAF1 91794 109 + 1 UAUGACCUAGUUAUUGCUAUUAGU----UUACCUUGACGCAGGCAUUCCAAAAUGGAUGCAUUACAUAGACACUAAGAAAACUCGUGUCUAUGCUGUUAUACUGUAAAGAAGA .......((((....))))....(----((((......((((((((((......))))))....(((((((((...........)))))))))))))......)))))..... ( -25.00) >DroPer_CAF1 2034 107 + 1 ----UUCUUUGGUGUGAAAAUAGAUCACUUACCUUAACACAAUAGUUCCAGAAUGGGUGCAUUAUAUAGACACUGACGAAACCAGUGUCUAUGCAGUUGUACUAUGG--GAAA ----......(((((((.......))))..)))............(((((.....((((((...(((((((((((.......)))))))))))....)))))).)))--)).. ( -30.70) >consensus UAUAUUCUUGGAAGCGUUAAUAGA____AUACCUUCACACAGUAGUUCCAAAAAGGGUGCAUCACAUAGACACUGAGGAAACUCGUGUCUAUACUGCUGUACUACGG__AAUA ......................................(((((.(((((.....)))........((((((((...........)))))))))).)))))............. (-13.80 = -13.05 + -0.75)
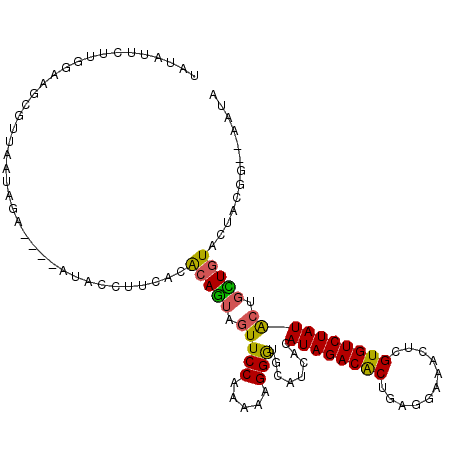
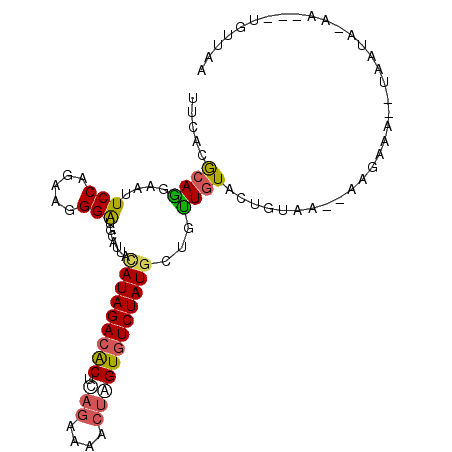
| Location | 7,788,399 – 7,788,493 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -13.05 |
| Energy contribution | -13.00 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7788399 94 + 22407834 UUCACACAGUAGUUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUAAA--UAAUAAGA---UAUUGA ......((((((((((......((((((.((.((((((((.(((....)))))))))))..)).))))))..))--)))...--........---))))). ( -25.70) >DroVir_CAF1 83812 98 + 1 UUCACGCAGGAAUUCCAGAACGGAUGCAUUAUAUAGACACUCAGAAAACUAGUGUCUAUGCUGUUGUACUGUCA--ACGAAACAUAAUA-AAUAAUGAAAA ((((....((....)).((.(((.((((.((((((((((((.((....)))))))))))).)).))))))))).--.............-.....)))).. ( -21.80) >DroWil_CAF1 91823 97 + 1 UUGACGCAGGCAUUCCAAAAUGGAUGCAUUACAUAGACACUAAGAAAACUCGUGUCUAUGCUGUUAUACUGUAAAGAAGAAC--AAAUA-AAUA-UGUAGG .....((((((((((......))))))....(((((((((...........)))))))))........))))..........--.....-....-...... ( -22.10) >DroMoj_CAF1 76743 97 + 1 UUUACGCAAGCAUUCCAGAAGGGAUGCAUAACAUAGACGCUCAGAAGGCUGGUAUCUAUGCUAUUGUACUGGAU--GAGAAAGAUAACA-GACA-UGUUAA .....(((((((((((....)))))))....((((((..(.(((....))))..))))))...)))).......--........(((((-....-))))). ( -25.70) >DroAna_CAF1 1280 91 + 1 UUUACGCAGUAAUUCCAGAAGGGGUGCAUAACAUAGACACUUAGCCAACUAGUGUCUAUGCUAUUGUACUGGAA--AAAA-----AACCACA---UACUCU .....(.((((...((....))((((((((.((((((((((.((....)))))))))))).)).))))))((..--....-----..))...---))))). ( -23.10) >DroPer_CAF1 2063 96 + 1 UUAACACAAUAGUUCCAGAAUGGGUGCAUUAUAUAGACACUGACGAAACCAGUGUCUAUGCAGUUGUACUAUGG--GAAAAAGAUAGUGAAA---UUUUCA ....(((.((..(((((.....((((((...(((((((((((.......)))))))))))....)))))).)))--)).....)).)))...---...... ( -28.70) >consensus UUCACGCAGGAAUUCCAGAAGGGAUGCAUUACAUAGACACUCAGAAAACUAGUGUCUAUGCUGUUGUACUGUAA__AAGAAA__UAAUA_AA___UGUUAA .....((((....(((.....))).......(((((((((.(((....))))))))))))...)))).................................. (-13.05 = -13.00 + -0.05)

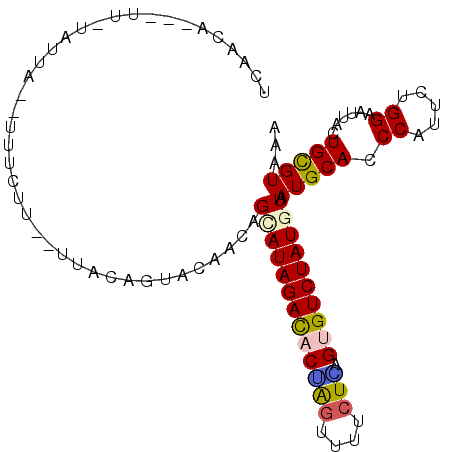
| Location | 7,788,399 – 7,788,493 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.53 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7788399 94 - 22407834 UCAAUA---UCUUAUUA--UUUAUU--CCGUAGUACAGCAGUAUAGACACGAGCUUCCUCAGCGUCUAUGUGAUGCACCCUUUUUGGAACUACUGUGUGAA ......---........--(((((.--(.(((((...(((.(((((((.((((....))).).)))))))...)))..((.....)).))))).).))))) ( -20.80) >DroVir_CAF1 83812 98 - 1 UUUUCAUUAUU-UAUUAUGUUUCGU--UGACAGUACAACAGCAUAGACACUAGUUUUCUGAGUGUCUAUAUAAUGCAUCCGUUCUGGAAUUCCUGCGUGAA ....((((((.-...........((--((......))))...(((((((((((....)).)))))))))))))))..(((((...((....)).))).)). ( -21.80) >DroWil_CAF1 91823 97 - 1 CCUACA-UAUU-UAUUU--GUUCUUCUUUACAGUAUAACAGCAUAGACACGAGUUUUCUUAGUGUCUAUGUAAUGCAUCCAUUUUGGAAUGCCUGCGUCAA ......-....-.....--...........(((.......(((((((((((((....))).))))))))))...((((((.....)).)))))))...... ( -22.30) >DroMoj_CAF1 76743 97 - 1 UUAACA-UGUC-UGUUAUCUUUCUC--AUCCAGUACAAUAGCAUAGAUACCAGCCUUCUGAGCGUCUAUGUUAUGCAUCCCUUCUGGAAUGCUUGCGUAAA .(((((-....-)))))........--......(((.((((((((((..((((....))).)..))))))))))((((.((....)).))))....))).. ( -23.40) >DroAna_CAF1 1280 91 - 1 AGAGUA---UGUGGUU-----UUUU--UUCCAGUACAAUAGCAUAGACACUAGUUGGCUAAGUGUCUAUGUUAUGCACCCCUUCUGGAAUUACUGCGUAAA ....((---((..((.-----....--((((((....((((((((((((((((....)).)))))))))))))).........))))))..))..)))).. ( -33.42) >DroPer_CAF1 2063 96 - 1 UGAAAA---UUUCACUAUCUUUUUC--CCAUAGUACAACUGCAUAGACACUGGUUUCGUCAGUGUCUAUAUAAUGCACCCAUUCUGGAACUAUUGUGUUAA .(((((---...........)))))--.(((((((....((((((((((((((.....)))))))))).....)))).((.....))...))))))).... ( -21.00) >consensus UCAACA___UU_UAUUA__UUUCUU__UUACAGUACAACAGCAUAGACACUAGUUUUCUCAGUGUCUAUGUAAUGCACCCAUUCUGGAAUUACUGCGUAAA ........................................(((((((((((((....))).)))))))))).(((((.((.....))......)))))... (-12.59 = -12.53 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:37 2006