
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,782,152 – 7,782,285 |
| Length | 133 |
| Max. P | 0.954205 |

| Location | 7,782,152 – 7,782,245 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.82 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -12.27 |
| Energy contribution | -10.08 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
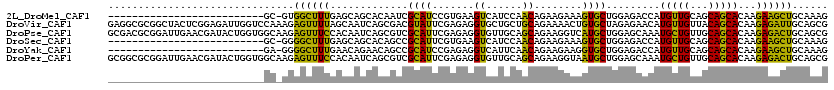
>2L_DroMel_CAF1 7782152 93 + 22407834 --------------------------GC-GUGGCUUUGAGCAGCACAAUCGCAUCCGUGAAGUCAUCCAACAGAAGAAAGUGCUGGAGACCAUGUUGCAGCAGCACAAGAAGCUGCAAAG --------------------------((-(((((((....((((((..((((....))))..((.((.....)).))..))))))))).))))))((((((..(....)..))))))... ( -29.90) >DroVir_CAF1 59566 120 + 1 GAGGCGCGGCUACUCGGAGAUUGGUCCAAAGAGUUUUAGCAAUCAGCGACGUAUUCGAGAGGUGCUGCUGCAGAAAACUGUGCUAGAGAACAUGUUGUUACAGCACAAGAGAUUGCAGCG .....(((.((.((((((......)))...((((....((.....)).....))))))))).))).(((((((.....((((((....(((.....)))..)))))).....))))))). ( -35.60) >DroPse_CAF1 33998 120 + 1 GCGACGCGGAUUGAACGAUACUGGUGGCAAGAGUUUCCACAAUCAGCGUCGCAUUCGAGAGGUGUUGCAGCAGAAGGUCAUGCUGGAGCAAAUGCUGUUGCAGCACAAGAGACUGCAGCG (((((((.(((((........(((..((....))..)))))))).)))))(((.((.....((((((((((((.......(((....)))....))))))))))))....)).))).)). ( -47.10) >DroSec_CAF1 35101 93 + 1 --------------------------GC-GGGGCUUUGAGCAGCACAGCCGCAUUCGUGAAGUCAUCCAACAGAAGAAAGUGCUGGAGACCAUGUUGCAGCAGCACAAGAAGCUGCAAAG --------------------------..-..((((.((.....)).))))(((..((((..(((.((((.((........)).))))))))))).))).(((((.......))))).... ( -29.00) >DroYak_CAF1 29934 93 + 1 --------------------------GA-GGGGCUUUGAACAGAACAGCCGCAUCCGAGAGGUCAUUCAACAGAAGAAGGUGCUGGAGACCAUGUUGCAGCAGCACAAGAAGCUGCAAAG --------------------------..-(.((((.(.......).)))).)........((((.((((.((........)).))))))))...(((((((..(....)..))))))).. ( -24.80) >DroPer_CAF1 34091 120 + 1 GCGGCGCGGAUUGAACGAUACUGGUGGCAAGAGUUUCCACAAUCAGCGUCGCAUUCGAGAGGUGUUGCAGCAGAAGGUAAUGCUGGAGCAAAUGCUGUUGCAGCACAAGAGACUGCAGCG (((((((.(((((........(((..((....))..)))))))).)))))(((.((.....((((((((((((.......(((....)))....))))))))))))....)).))).)). ( -46.50) >consensus __________________________GC_AGAGCUUUGACAAGCACAGUCGCAUUCGAGAGGUCAUGCAACAGAAGAAAGUGCUGGAGAACAUGUUGCAGCAGCACAAGAAACUGCAAAG ...............................((((((.............((((.......((......))........)))).........(((((...)))))...))))))...... (-12.27 = -10.08 + -2.19)

| Location | 7,782,152 – 7,782,245 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.82 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -13.04 |
| Energy contribution | -11.79 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7782152 93 - 22407834 CUUUGCAGCUUCUUGUGCUGCUGCAACAUGGUCUCCAGCACUUUCUUCUGUUGGAUGACUUCACGGAUGCGAUUGUGCUGCUCAAAGCCAC-GC-------------------------- ..(((((((..(....)..)))))))..((((...((((((..((.(((((.(((....))))))))...))..))))))......)))).-..-------------------------- ( -29.30) >DroVir_CAF1 59566 120 - 1 CGCUGCAAUCUCUUGUGCUGUAACAACAUGUUCUCUAGCACAGUUUUCUGCAGCAGCACCUCUCGAAUACGUCGCUGAUUGCUAAAACUCUUUGGACCAAUCUCCGAGUAGCCGCGCCUC .((((((.....((((((((.(((.....)))...)))))))).....)))))).........((..(((.(((..((((((((((....)))))..)))))..))))))..))...... ( -32.00) >DroPse_CAF1 33998 120 - 1 CGCUGCAGUCUCUUGUGCUGCAACAGCAUUUGCUCCAGCAUGACCUUCUGCUGCAACACCUCUCGAAUGCGACGCUGAUUGUGGAAACUCUUGCCACCAGUAUCGUUCAAUCCGCGUCGC .((.((((....(..(((((....(((....))).)))))..)....)))).))..............(((((((.(((((..((.(((.........))).))...))))).))))))) ( -39.60) >DroSec_CAF1 35101 93 - 1 CUUUGCAGCUUCUUGUGCUGCUGCAACAUGGUCUCCAGCACUUUCUUCUGUUGGAUGACUUCACGAAUGCGGCUGUGCUGCUCAAAGCCCC-GC-------------------------- (((((.(((.....(..(.((((((....(((((((((((........))))))).)))).......)))))).)..).))))))))....-..-------------------------- ( -32.60) >DroYak_CAF1 29934 93 - 1 CUUUGCAGCUUCUUGUGCUGCUGCAACAUGGUCUCCAGCACCUUCUUCUGUUGAAUGACCUCUCGGAUGCGGCUGUUCUGUUCAAAGCCCC-UC-------------------------- ....(((((.......))))).(((.(..((((..(((((........)))))...)))).....).)))((((...........))))..-..-------------------------- ( -25.40) >DroPer_CAF1 34091 120 - 1 CGCUGCAGUCUCUUGUGCUGCAACAGCAUUUGCUCCAGCAUUACCUUCUGCUGCAACACCUCUCGAAUGCGACGCUGAUUGUGGAAACUCUUGCCACCAGUAUCGUUCAAUCCGCGCCGC .((.((((......((((((....(((....))).))))))......)))).))..............(((.(((.(((((..((.(((.........))).))...))))).))).))) ( -33.80) >consensus CGCUGCAGCCUCUUGUGCUGCAACAACAUGGUCUCCAGCACUUUCUUCUGCUGCAUCACCUCUCGAAUGCGACGCUGAUGGUCAAAACCCC_GC__________________________ .((.((((......((((((...............))))))......)))).)).................................................................. (-13.04 = -11.79 + -1.25)

| Location | 7,782,165 – 7,782,285 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -20.98 |
| Energy contribution | -23.76 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
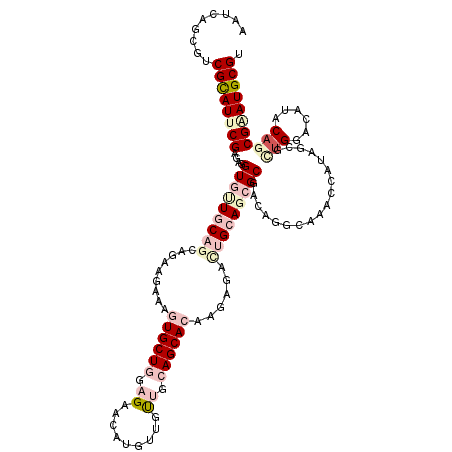
>2L_DroMel_CAF1 7782165 120 + 22407834 CAGCACAAUCGCAUCCGUGAAGUCAUCCAACAGAAGAAAGUGCUGGAGACCAUGUUGCAGCAGCACAAGAAGCUGCAAAGCGAUCGACACACCAUAGCGAUGGAUAUCCAGCGAAUGCGU ..((((.(((((...((((..(((.((((.((........)).)))))))))))(((((((..(....)..))))))).))))).)...........((.(((....))).))..))).. ( -34.00) >DroVir_CAF1 59606 120 + 1 AAUCAGCGACGUAUUCGAGAGGUGCUGCUGCAGAAAACUGUGCUAGAGAACAUGUUGUUACAGCACAAGAGAUUGCAGCGCGACAGGCAAACGAUUGCCUCGGACAUACAGCGGAUGCGU ........(((((((((....((((((((((((.....((((((....(((.....)))..)))))).....))))))).....((((((....)))))).)).)))....))))))))) ( -40.40) >DroPse_CAF1 34038 120 + 1 AAUCAGCGUCGCAUUCGAGAGGUGUUGCAGCAGAAGGUCAUGCUGGAGCAAAUGCUGUUGCAGCACAAGAGACUGCAGCGCGACAGGCAGACCAUAGCUCUGGACAUACAGCGGAUGCGU .........((((((((....(((((.(((.((..((((.((((.((((....)))((((((((......).)))))))....).))))))))....))))))))))....)))))))). ( -44.00) >DroYak_CAF1 29947 120 + 1 CAGAACAGCCGCAUCCGAGAGGUCAUUCAACAGAAGAAGGUGCUGGAGACCAUGUUGCAGCAGCACAAGAAGCUGCAAAGCGAUCGUCACACCAUAGCAAUGGACAUCCAGCGAAUGCGU .........(((((((....))..................((((((((((...(((((.(((((.......)))))...))))).)))...((((....))))...))))))).))))). ( -36.50) >DroMoj_CAF1 42299 120 + 1 AAUCAGCGCCGCAUUCGUGAGGUGUUGCUGCAAAAAACUGUGCUAGAGAAUAUGUUGUUACAGCACAAGAGAUUGCAGCGCGACAGGCAAACAAUUGCGCUGGAUAUACAGCGAAUGCGA .........(((((((((...((((((((((((.....((((((.((.((....)).))..)))))).....)))))))....(((((((....)))).))))))))...))))))))). ( -42.70) >DroPer_CAF1 34131 120 + 1 AAUCAGCGUCGCAUUCGAGAGGUGUUGCAGCAGAAGGUAAUGCUGGAGCAAAUGCUGUUGCAGCACAAGAGACUGCAGCGCGACAGGCAGACCAUAGCUCUGGACAUACAGCGGAUGCGU .........((((((((....(((((.(((((........)))))((((...(((.((((((((......).))))))))))...((....))...))))..)))))....)))))))). ( -43.40) >consensus AAUCAGCGUCGCAUUCGAGAGGUGUUGCAGCAGAAGAAAGUGCUGGAGAACAUGUUGUUGCAGCACAAGAGACUGCAGCGCGACAGGCAAACCAUAGCGCUGGACAUACAGCGAAUGCGU .........((((((((....(((((((((.........((((((.((.........)).))))))......)))))))))..................(((......))))))))))). (-20.98 = -23.76 + 2.78)

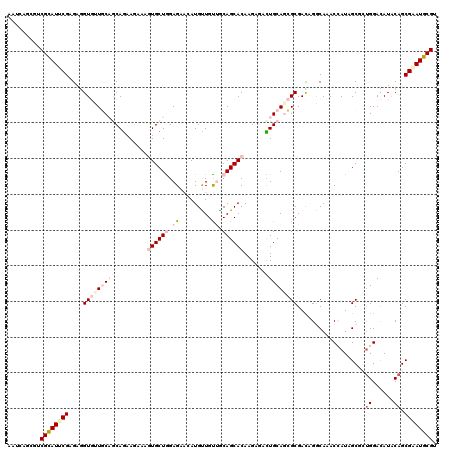
| Location | 7,782,165 – 7,782,285 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -38.51 |
| Consensus MFE | -21.28 |
| Energy contribution | -19.81 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
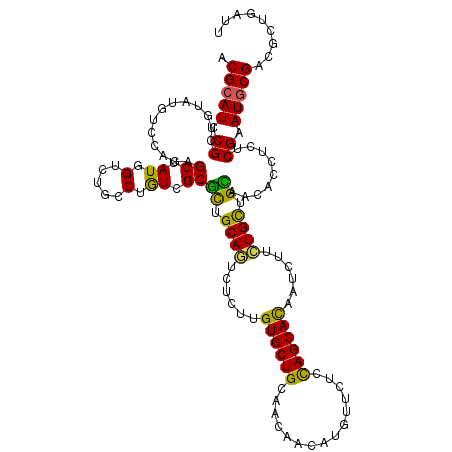
>2L_DroMel_CAF1 7782165 120 - 22407834 ACGCAUUCGCUGGAUAUCCAUCGCUAUGGUGUGUCGAUCGCUUUGCAGCUUCUUGUGCUGCUGCAACAUGGUCUCCAGCACUUUCUUCUGUUGGAUGACUUCACGGAUGCGAUUGUGCUG .((((((((((((....)))..))....(((.((((((((..(((((((..(....)..)))))))..)))))(((((((........))))))).)))..))))))))))......... ( -44.70) >DroVir_CAF1 59606 120 - 1 ACGCAUCCGCUGUAUGUCCGAGGCAAUCGUUUGCCUGUCGCGCUGCAAUCUCUUGUGCUGUAACAACAUGUUCUCUAGCACAGUUUUCUGCAGCAGCACCUCUCGAAUACGUCGCUGAUU ....(((.(((((((...((((((((....)))))....((((((((.....((((((((.(((.....)))...)))))))).....)))))).)).....))).)))))..)).))). ( -35.90) >DroPse_CAF1 34038 120 - 1 ACGCAUCCGCUGUAUGUCCAGAGCUAUGGUCUGCCUGUCGCGCUGCAGUCUCUUGUGCUGCAACAGCAUUUGCUCCAGCAUGACCUUCUGCUGCAACACCUCUCGAAUGCGACGCUGAUU ....(((.((.(((.(.(((......))).))))..(((((((.((((....(..(((((....(((....))).)))))..)....)))).)).....(....)...))))))).))). ( -34.30) >DroYak_CAF1 29947 120 - 1 ACGCAUUCGCUGGAUGUCCAUUGCUAUGGUGUGACGAUCGCUUUGCAGCUUCUUGUGCUGCUGCAACAUGGUCUCCAGCACCUUCUUCUGUUGAAUGACCUCUCGGAUGCGGCUGUUCUG .(((((((((((((.(((((((.....)))).)))(((((..(((((((..(....)..)))))))..)))))))))))....(((((....))).))......)))))))......... ( -43.50) >DroMoj_CAF1 42299 120 - 1 UCGCAUUCGCUGUAUAUCCAGCGCAAUUGUUUGCCUGUCGCGCUGCAAUCUCUUGUGCUGUAACAACAUAUUCUCUAGCACAGUUUUUUGCAGCAACACCUCACGAAUGCGGCGCUGAUU (((((((((.((......(((.((((....)))))))....(((((((....((((((((...............))))))))....))))))).......)))))))))))........ ( -39.16) >DroPer_CAF1 34131 120 - 1 ACGCAUCCGCUGUAUGUCCAGAGCUAUGGUCUGCCUGUCGCGCUGCAGUCUCUUGUGCUGCAACAGCAUUUGCUCCAGCAUUACCUUCUGCUGCAACACCUCUCGAAUGCGACGCUGAUU ....(((.((.(((.(.(((......))).))))..(((((((.((((......((((((....(((....))).))))))......)))).)).....(....)...))))))).))). ( -33.50) >consensus ACGCAUCCGCUGUAUGUCCAGAGCUAUGGUCUGCCUGUCGCGCUGCAGUCUCUUGUGCUGCAACAACAUGUUCUCCAGCACAAUCUUCUGCUGCAACACCUCUCGAAUGCGACGCUGAUU .(((((.((.............((.((.(.....).)).))((.((((......((((((...............))))))......)))).)).........)).)))))......... (-21.28 = -19.81 + -1.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:33 2006