
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,780,466 – 7,780,581 |
| Length | 115 |
| Max. P | 0.798332 |

| Location | 7,780,466 – 7,780,581 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -26.83 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
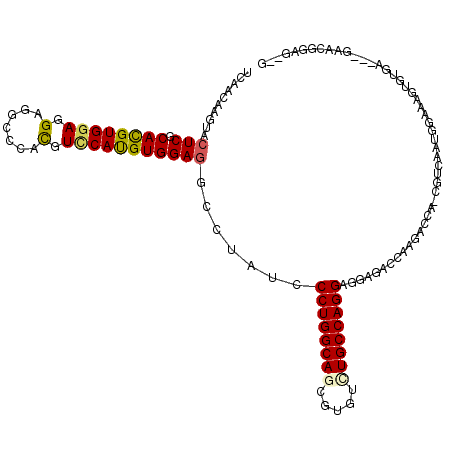
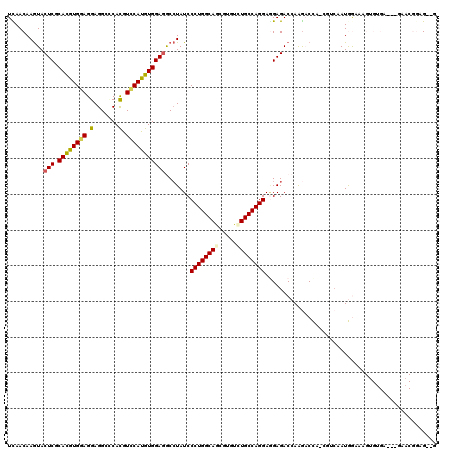
>2L_DroMel_CAF1 7780466 115 + 22407834 UUAACAAAUACUCGCACGUGGAGGAGGCGCACGUUCAUGUGGAGGCGUAUCCCUGGCAGCGAGUUUGCCAGGAGGAGACCAGGACCAACGUCAAUGGGAAGUGCGA---GAACGGAG--U ..........(((((((.(((.(((.((((.(.(((....)))))))).)))(((((((.....))))))).......))).(((....)))........))))))---).......--. ( -37.90) >DroPse_CAF1 32281 100 + 1 UCAACAAGUACUCGCAUGUGGAGGAGGCCCACGUCCAUGUGGAGACCUAUCCCUGGCAGCGUGUCUGCCAGGAGGAGACAAAGUCU---GUCAAUAAUCA---------------GG--G ..........(((.((((((((.(.......).))))))))))).(((.((((((((((.....)))))))).)).((((.....)---))).......)---------------))--. ( -35.70) >DroEre_CAF1 27989 115 + 1 UCAACAAGUACUCGCACGUGGAGGAGGCGCACGUCCAUGUGGAGGCCUAUCCCUGGCAGCGAGUAUGCCAGGAGGAGACCAGGACCACCCUCAAUGGAAAGUGUGA---GAACGGAG--U .......((.(((((((((((.(((.(....).))).(.(((...(((...(((((((.......))))))))))...))).).))))((.....))...))))))---).))....--. ( -42.60) >DroWil_CAF1 42499 120 + 1 UCAACAAGUACUCGCAUGUGGAGGAGGCUCAUGUGCACGUGGAAGCCUAUCCCUGGCAACGUGUGUGCCAGGAGGAGACCAAGACCAUGUGGAAUGGAAAGGGUGCAUGCAACGGCGAUC .......(((((((((((((.((....)))))))))...(((...(((...(((((((.......))))))))))...)))...((((.....))))...)))))).(((....)))... ( -39.20) >DroYak_CAF1 28251 115 + 1 UUAACAAGUACUCGCACGUGGAGGAGGCCCACGUCCAUGUGGAGGCCUAUCCCUGGCAGCGUGUUUGCCAGGAAGAGACCAGAACCCCCAGCAAUGGAAAGUGUGA---GAACGGUG--G ....((.((.(((((((.(((...(((((....(((....))))))))...((((((((.....))))))))......)))......(((....)))...))))))---).))..))--. ( -43.90) >DroPer_CAF1 32374 100 + 1 UCAACAAGUACUCGCAUGUGGAGGAGGCCCACGUCCAUGUGGAGACCUAUCCCUGGCAGCGUGUCUGCCAGGAUGAGACAAAGUCU---GUCAAUAAUCA---------------GG--G ..........(((.((((((((.(.......).))))))))))).(((...((((((((.....))))))))....((((.....)---))).......)---------------))--. ( -35.50) >consensus UCAACAAGUACUCGCACGUGGAGGAGGCCCACGUCCAUGUGGAGGCCUAUCCCUGGCAGCGUGUCUGCCAGGAGGAGACCAAGACCA_CGUCAAUGGAAAGUGUGA___GAACGGAG__G ..........(((.((((((((.(.......).))))))))))).......((((((((.....))))))))................................................ (-26.83 = -26.75 + -0.08)

| Location | 7,780,466 – 7,780,581 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
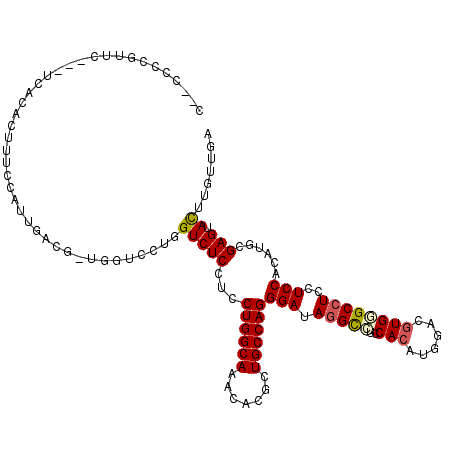
>2L_DroMel_CAF1 7780466 115 - 22407834 A--CUCCGUUC---UCGCACUUCCCAUUGACGUUGGUCCUGGUCUCCUCCUGGCAAACUCGCUGCCAGGGAUACGCCUCCACAUGAACGUGCGCCUCCUCCACGUGCGAGUAUUUGUUAA .--.......(---((((((..........((((((....(((....((((((((.......))))))))....))).))).)))...(((.(......))))))))))).......... ( -32.60) >DroPse_CAF1 32281 100 - 1 C--CC---------------UGAUUAUUGAC---AGACUUUGUCUCCUCCUGGCAGACACGCUGCCAGGGAUAGGUCUCCACAUGGACGUGGGCCUCCUCCACAUGCGAGUACUUGUUGA .--..---------------........(((---((...)))))....((((((((.....))))))))((((((((((((..((((...((....))))))..)).))).))))))).. ( -32.40) >DroEre_CAF1 27989 115 - 1 A--CUCCGUUC---UCACACUUUCCAUUGAGGGUGGUCCUGGUCUCCUCCUGGCAUACUCGCUGCCAGGGAUAGGCCUCCACAUGGACGUGCGCCUCCUCCACGUGCGAGUACUUGUUGA .--....((.(---((.(((((((....)))))))((((.(((((..((((((((.......))))))))..))))).......))))(..((.........))..)))).))....... ( -40.00) >DroWil_CAF1 42499 120 - 1 GAUCGCCGUUGCAUGCACCCUUUCCAUUCCACAUGGUCUUGGUCUCCUCCUGGCACACACGUUGCCAGGGAUAGGCUUCCACGUGCACAUGAGCCUCCUCCACAUGCGAGUACUUGUUGA ........(((((((..............(((.(((....(((((..((((((((.......))))))))..))))).))).))).....(((....)))..)))))))........... ( -33.30) >DroYak_CAF1 28251 115 - 1 C--CACCGUUC---UCACACUUUCCAUUGCUGGGGGUUCUGGUCUCUUCCUGGCAAACACGCUGCCAGGGAUAGGCCUCCACAUGGACGUGGGCCUCCUCCACGUGCGAGUACUUGUUAA .--....((.(---((.(((..(((((.(.(((((((.(((.....(((((((((.......))))))))))))))))))))))))).(((((.....)))))))).))).))....... ( -44.50) >DroPer_CAF1 32374 100 - 1 C--CC---------------UGAUUAUUGAC---AGACUUUGUCUCAUCCUGGCAGACACGCUGCCAGGGAUAGGUCUCCACAUGGACGUGGGCCUCCUCCACAUGCGAGUACUUGUUGA .--..---------------........(((---((...)))))....((((((((.....))))))))((((((((((((..((((...((....))))))..)).))).))))))).. ( -32.40) >consensus C__CCCCGUUC___UCACACUUUCCAUUGACG_UGGUCCUGGUCUCCUCCUGGCAAACACGCUGCCAGGGAUAGGCCUCCACAUGGACGUGGGCCUCCUCCACAUGCGAGUACUUGUUGA .........................................(((((...((((((.......))))))(((.(((((..(((......))))))))..)))......))).))....... (-21.53 = -21.67 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:29 2006