
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,730,400 – 7,730,555 |
| Length | 155 |
| Max. P | 0.785925 |

| Location | 7,730,400 – 7,730,515 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.77 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -16.41 |
| Energy contribution | -18.20 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7730400 115 + 22407834 CGGGCGGCUAGGGCCAGACUACCGCCCAUUCCGCCUCCGGACUCCGUUUCCACUGGCUCCCCGAUAAAAU-CACCGGGCAG-UUCUGUGCCAUGUUCACUAUUCUCACAGUCGCCCA .((((((((..(((((((..((.((((.....(((...(((.......)))...))).....(((...))-)...)))).)-))))).)))..(....).........)))))))). ( -40.00) >DroSim_CAF1 21347 115 + 1 CGGGCGGCUAGGGCCAGACUACCGCCCAUUCCGCCUCCGGACUCCGUUUCCACGGGCUCUCCGGUAAAAU-CACCGGGCAG-CUCUGUGCCAUGUUCACUAUUCUCACAGUCGCCCA .((((((((.((((.........))))..((((....)))).........((((((((.((((((.....-.)))))).))-).)))))...................)))))))). ( -45.20) >DroPer_CAF1 14066 109 + 1 CGCGUUGAGGCAGCCAAUCCUCC---UCCUCCGCCUCCGC-CUCC---UCCAACAGCAACUCCAUCUUCUACUCCA-GCUGCUGCGGCUUCAUCUCCACCGUUUUCGCAGUCGCCCA .(((..(((((............---......))))))))-....---............................-((.(((((((.................))))))).))... ( -20.10) >consensus CGGGCGGCUAGGGCCAGACUACCGCCCAUUCCGCCUCCGGACUCCGUUUCCACCGGCUCCCCGAUAAAAU_CACCGGGCAG_UUCUGUGCCAUGUUCACUAUUCUCACAGUCGCCCA .((((((((((((((..............((((....)))).............))))))...............(.((((...)))).)..................)))))))). (-16.41 = -18.20 + 1.79)



| Location | 7,730,400 – 7,730,515 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.77 |
| Mean single sequence MFE | -45.26 |
| Consensus MFE | -21.25 |
| Energy contribution | -24.90 |
| Covariance contribution | 3.65 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7730400 115 - 22407834 UGGGCGACUGUGAGAAUAGUGAACAUGGCACAGAA-CUGCCCGGUG-AUUUUAUCGGGGAGCCAGUGGAAACGGAGUCCGGAGGCGGAAUGGGCGGUAGUCUGGCCCUAGCCGCCCG .((((((((((....)))))......(((.(((((-(((((((...-......((.(((..((.((....))))..))).)).......))))))))..))))))).....))))). ( -48.19) >DroSim_CAF1 21347 115 - 1 UGGGCGACUGUGAGAAUAGUGAACAUGGCACAGAG-CUGCCCGGUG-AUUUUACCGGAGAGCCCGUGGAAACGGAGUCCGGAGGCGGAAUGGGCGGUAGUCUGGCCCUAGCCGCCCG .((((((((((....)))))......(((.((((.-((((((((((-....)))))....(((((((....)....((((....)))))))))))))))))))))).....))))). ( -53.10) >DroPer_CAF1 14066 109 - 1 UGGGCGACUGCGAAAACGGUGGAGAUGAAGCCGCAGCAGC-UGGAGUAGAAGAUGGAGUUGCUGUUGGA---GGAG-GCGGAGGCGGAGGA---GGAGGAUUGGCUGCCUCAACGCG ...(((.((.(((...((((.........))))(((((((-(..(........)..))))))))))).)---)...-...(((((((..((---......))..)))))))..))). ( -34.50) >consensus UGGGCGACUGUGAGAAUAGUGAACAUGGCACAGAA_CUGCCCGGUG_AUUUUAUCGGGGAGCCAGUGGAAACGGAGUCCGGAGGCGGAAUGGGCGGUAGUCUGGCCCUAGCCGCCCG .((((((((((....)))))......(((.((((..((((((((((.....)))).....(((..((((.......))))..))).....))))))...))))))).....))))). (-21.25 = -24.90 + 3.65)

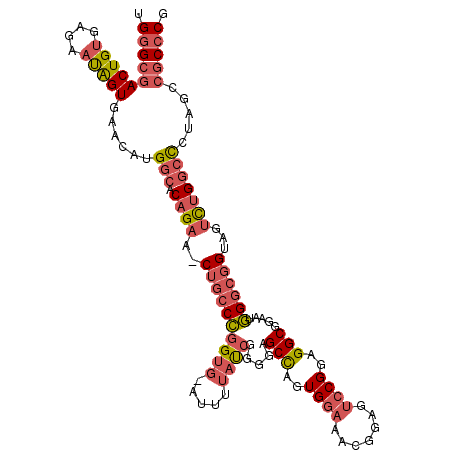
| Location | 7,730,437 – 7,730,555 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -20.34 |
| Energy contribution | -25.03 |
| Covariance contribution | 4.69 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7730437 118 - 22407834 AAGGGCAUCUGCGGUGAGCAGCUCAACAAUUGCAUGCUCCUGGGCGACUGUGAGAAUAGUGAACAUGGCACAGAA-CUGCCCGGUG-AUUUUAUCGGGGAGCCAGUGGAAACGGAGUCCG ..((((.(((((((.(((((((.........)).))))))))(((..(((((...((.(....)))..)))))..-...(((((((-....)))))))..)))........)))))))). ( -42.80) >DroPse_CAF1 14184 115 - 1 AAAGGCAUCUGCGGGGAGCAGCUCAACAACUGCAUGCUGCUGGGCGACUGCGAAAACGGUGGAGAUGGAGCCGCAGCAGU-UGGAGUAGAAGAUGGAGUUGCUGUUGGA---GGAG-GCG ....((.(((.(.((.((((((((..(..((((..((((((((((..(..................)..))).)))))))-....))))..)...)))))))).)).).---))).-)). ( -42.47) >DroSim_CAF1 21384 118 - 1 AAGGGCAUCUGCGGUGAGCAGCUCAACAAUUGCAUGCUCCUGGGCGACUGUGAGAAUAGUGAACAUGGCACAGAG-CUGCCCGGUG-AUUUUACCGGAGAGCCCGUGGAAACGGAGUCCG ..((((.(((.(((((((((((.........)).)))))(((((((.(((((...((.(....)))..)))))..-.)))))))..-.....)))).)))..((((....)))).)))). ( -48.30) >DroPer_CAF1 14100 115 - 1 AAAGGCAUCUGCGGGGAGCAGCUCAACAACUGCAUGCUGCUGGGCGACUGCGAAAACGGUGGAGAUGAAGCCGCAGCAGC-UGGAGUAGAAGAUGGAGUUGCUGUUGGA---GGAG-GCG ....((.(((.(.((.((((((((..(..((((..((((((((((.((((......)))).........))).)))))))-....))))..)...)))))))).)).).---))).-)). ( -43.80) >consensus AAAGGCAUCUGCGGGGAGCAGCUCAACAACUGCAUGCUCCUGGGCGACUGCGAAAACAGUGAACAUGGAACAGAA_CAGC_CGGAG_AGAAGAUCGAGGAGCCGGUGGA___GGAG_CCG .....(((((.(.(((((((((.........)).))))))).)...((((......))))..)))))...((((((((((((((((.....))))))))))))))))............. (-20.34 = -25.03 + 4.69)

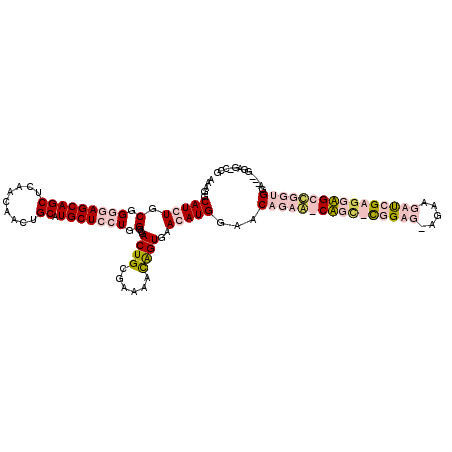

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:19 2006