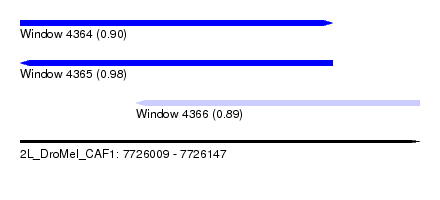
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,726,009 – 7,726,147 |
| Length | 138 |
| Max. P | 0.981213 |

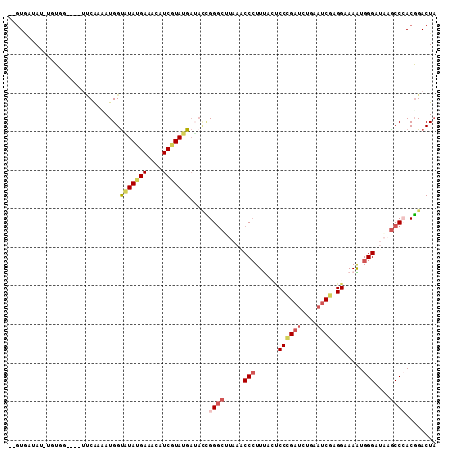
| Location | 7,726,009 – 7,726,117 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7726009 108 + 22407834 AAGUCCGUUGGCUUAUCCCAUUUUCCUCGAUUCAGAUCGGGGGAAAAGGGUUUAUGCCCGGUAUCAUACGAUGUUUCAUAUACCAUGUUGAACUACCCACA-AUAUUAC-- ....(((..(((....(((.(((((((((((....))))))))))).))).....))))))((((....))))...........((((((.........))-))))...-- ( -28.40) >DroGri_CAF1 6715 103 + 1 UAGUCCGUUGGCUGUUCCCAUUUGCCACGAUUCAAGUCUGGUUUGAAUGGCCUGCGGCCACUCUCAUAGGAAGUCUCAUAUACCAUUGCGGU------A-U-AUAGUACUC ...(((..(((((((..((((((((((.(((....)))))))..))))))...)))))))........)))(((((.(((((((.....)))------)-)-)))).))). ( -32.10) >DroSec_CAF1 14549 105 + 1 UAGUCCGUGGGCUUAUCCCACUUUCCUCGAUUUAAAUCGGGAGUAAAGGGUUUAAGCCCGGUAUCAUACGAUGUUUCAUACACCAUAUUGAA---CCCACA-AUAUCAC-- ........(((((((..((.(((((((((((....)))))).).))))))..)))))))((((((....)))((.....)))))((((((..---....))-))))...-- ( -29.60) >DroSim_CAF1 16947 105 + 1 UAGUCCGUGGGCUUAUCCCACUUUCCUCGAUUUAGAUCGGGAGUAAAGGGUUUAAGCCCGGUAUCAUACGAUGUUUCAUACACCAUGUUGAA---CCCACA-AUAUCAC-- ......(((((((((..((.(((((((((((....)))))).).))))))..)))))))((..(((.(((.(((.....)))...)))))).---.)))).-.......-- ( -29.90) >DroWil_CAF1 6615 107 + 1 UAGUCAGUGGGUUUUUCCCAUUUGCCACGUUGCAGAUCGGGACGAAAGGGCUUUUGCCCCGAUUCGUAUGAAGAUUCAUAAACCAUUUCGGC----GUUGGAAUUGGAAUG ......(((((((((((((((((((......)))))).)))(((((.((((....))))...)))))..))))))))))...(((.(((...----....))).))).... ( -33.00) >DroYak_CAF1 9290 104 + 1 UAGUCCGUGGGCUUAUCCCACUUGCCACGAUUCAGAUCAGGGGAGAAGGGCUUAAGCCCAAUAGCAUACGAUGUUUCAUAUGCCAU--UGUA----UUAGA-AUAUCACUC ..((...((((((((.(((.(((.((..(((....))).)).)))..)))..))))))))...))....(((((((...((((...--.)))----)..))-))))).... ( -31.50) >consensus UAGUCCGUGGGCUUAUCCCACUUGCCACGAUUCAGAUCGGGAGUAAAGGGCUUAAGCCCGGUAUCAUACGAUGUUUCAUAUACCAUGUUGAA____CCACA_AUAUCAC__ ......(((((.....)))))..(((.((((....))))))).....((((....)))).................................................... (-11.26 = -11.90 + 0.64)

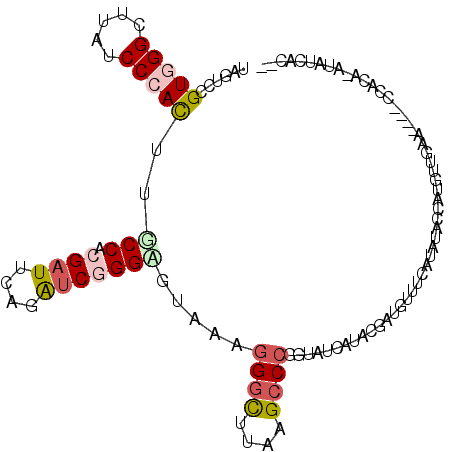
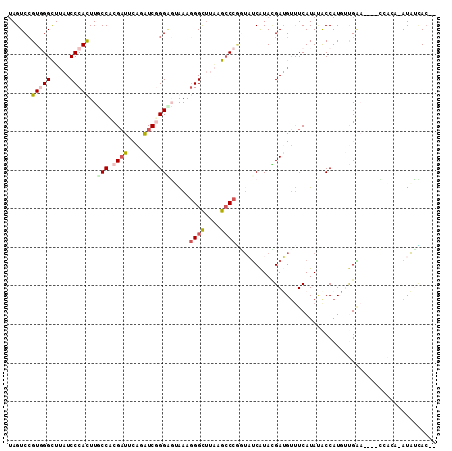
| Location | 7,726,009 – 7,726,117 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -12.34 |
| Energy contribution | -13.48 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7726009 108 - 22407834 --GUAAUAU-UGUGGGUAGUUCAACAUGGUAUAUGAAACAUCGUAUGAUACCGGGCAUAAACCCUUUUCCCCCGAUCUGAAUCGAGGAAAAUGGGAUAAGCCAACGGACUU --.......-.......(((((....((((((((((....)))))...)))))(((.((..((.((((((..((((....)))).)))))).))..)).)))...))))). ( -28.00) >DroGri_CAF1 6715 103 - 1 GAGUACUAU-A-U------ACCGCAAUGGUAUAUGAGACUUCCUAUGAGAGUGGCCGCAGGCCAUUCAAACCAGACUUGAAUCGUGGCAAAUGGGAACAGCCAACGGACUA ((((..(((-(-(------(((.....))))))))..)))).......((((((((...))))))))..............(((((((...((....))))).)))).... ( -31.50) >DroSec_CAF1 14549 105 - 1 --GUGAUAU-UGUGGG---UUCAAUAUGGUGUAUGAAACAUCGUAUGAUACCGGGCUUAAACCCUUUACUCCCGAUUUAAAUCGAGGAAAGUGGGAUAAGCCCACGGACUA --......(-(((.((---(...(((((((((.....)))))))))...)))(((((((..(((((...(((((((....)))).)))))).))..))))))))))).... ( -37.10) >DroSim_CAF1 16947 105 - 1 --GUGAUAU-UGUGGG---UUCAACAUGGUGUAUGAAACAUCGUAUGAUACCGGGCUUAAACCCUUUACUCCCGAUCUAAAUCGAGGAAAGUGGGAUAAGCCCACGGACUA --.......-....((---(((.....(((((((((....))))))...)))(((((((..(((((...(((((((....)))).)))))).))..)))))))..))))). ( -34.20) >DroWil_CAF1 6615 107 - 1 CAUUCCAAUUCCAAC----GCCGAAAUGGUUUAUGAAUCUUCAUACGAAUCGGGGCAAAAGCCCUUUCGUCCCGAUCUGCAACGUGGCAAAUGGGAAAAACCCACUGACUA ...............----((((...((...(((((....)))))((.((((((((.(((....))).)))))))).))))...))))...((((.....))))....... ( -26.50) >DroYak_CAF1 9290 104 - 1 GAGUGAUAU-UCUAA----UACA--AUGGCAUAUGAAACAUCGUAUGCUAUUGGGCUUAAGCCCUUCUCCCCUGAUCUGAAUCGUGGCAAGUGGGAUAAGCCCACGGACUA ..(((....-.....----..((--(((((((((((....)))))))))))))((((((..(((..((..((((((....)))).))..)).))).)))))))))...... ( -36.40) >consensus __GUGAUAU_UGUGG____UUCAAAAUGGUAUAUGAAACAUCGUAUGAUACCGGGCUUAAACCCUUUACUCCCGAUCUGAAUCGAGGAAAAUGGGAUAAGCCCACGGACUA .............................(((((((....))))))).....((((.....(((......((((((....)))).)).....)))....))))........ (-12.34 = -13.48 + 1.14)

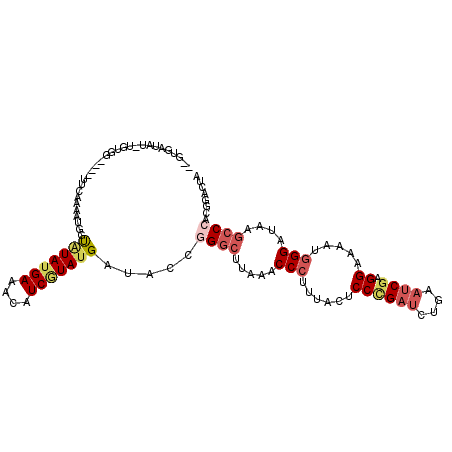
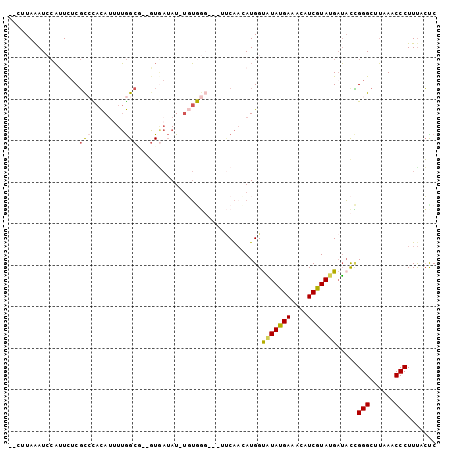
| Location | 7,726,049 – 7,726,147 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 68.69 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -8.12 |
| Energy contribution | -8.20 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7726049 98 - 22407834 --CUUAAAUCCAUUCUCGCCCACAUUUUGGCG--GUAAUAU-UGUGGGUAGUUCAACAUGGUAUAUGAAACAUCGUAUGAUACCGGGCAUAAACCCUUUUCCC --.....((((((..(((((........))))--)......-.))))))..........(((((((((....)))))...))))(((......)))....... ( -19.70) >DroSec_CAF1 14589 95 - 1 --UUUAAAUCCAUUCUCGCCCACAUUUUGGCG--GUGAUAU-UGUGGG---UUCAAUAUGGUGUAUGAAACAUCGUAUGAUACCGGGCUUAAACCCUUUACUC --(((((..(((((..((((........))))--..)))..-....((---(...(((((((((.....)))))))))...)))))..))))).......... ( -25.50) >DroSim_CAF1 16987 95 - 1 --GUUAAAUCCAUUCUCGCCCACAUUUUGGCU--GUGAUAU-UGUGGG---UUCAACAUGGUGUAUGAAACAUCGUAUGAUACCGGGCUUAAACCCUUUACUC --(((.(((((((..(((((((.....)))..--))))...-.)))))---)).)))..(((((((((....))))))...)))(((......)))....... ( -22.90) >DroWil_CAF1 6655 99 - 1 GACAUAGAUAAUCUUACCUAUCUAUAUAAGCACAUUCCAAUUCCAAC----GCCGAAAUGGUUUAUGAAUCUUCAUACGAAUCGGGGCAAAAGCCCUUUCGUC ...(((((((........))))))).......(((.(((.(((....----...))).)))...))).........(((((..(((((....)))))))))). ( -21.40) >DroYak_CAF1 9330 94 - 1 --UUUAAAUACACUUUCGUGUACAAAUUGGCUGAGUGAUAU-UCUAA----UACA--AUGGCAUAUGAAACAUCGUAUGCUAUUGGGCUUAAGCCCUUCUCCC --......(((((....)))))......(((((((((....-.)...----..((--(((((((((((....))))))))))))).)))).))))........ ( -27.10) >consensus __CUUAAAUCCAUUCUCGCCCACAUUUUGGCG__GUGAUAU_UGUGGG___UUCAACAUGGUAUAUGAAACAUCGUAUGAUACCGGGCUUAAACCCUUUACUC .................(((........)))..............................(((((((....))))))).....(((......)))....... ( -8.12 = -8.20 + 0.08)

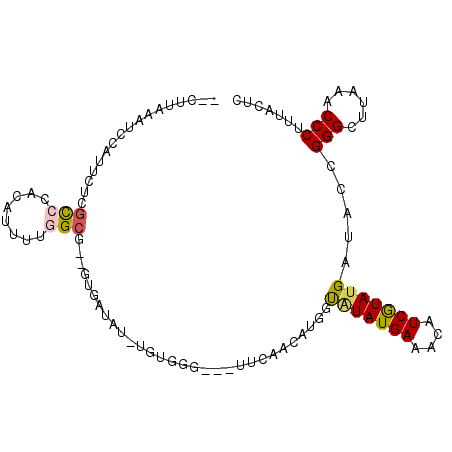
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:13 2006