
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,714,112 – 7,714,392 |
| Length | 280 |
| Max. P | 0.896363 |

| Location | 7,714,112 – 7,714,232 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -27.29 |
| Energy contribution | -28.60 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7714112 120 - 22407834 GGAGCGUCUAAAGCGCUCCUGCCGACCCACAGAUUGGUAUCCGGUUGAGAUGAAACAUCGUCAGCAUUACGGGGAUCGUGUCGGAAAUAUUGAGAUGACCGACCAUCUUUUCAAUGGUAU (((((((.....)))))))(((((......((((.(((..((((((((((((...)))).)))))....)))((.((((.((((.....)))).)))))).)))))))......))))). ( -38.40) >DroSec_CAF1 3105 120 - 1 GGAGCGUCUAAAACGCUCCUGCCGACCCACAGAUUGGUAUCCGGUUGAUGUGAAACAUCCUCAGCAUUACGGGGAUCUCGUCGGAAACAUUGAGAUGACAGACCAUCUUUCUAAUGGUAC (((((((.....)))))))..(((((.((((((((((...))))))..))))....((((((........))))))...)))))...((((((((((......))))))...)))).... ( -38.90) >DroSim_CAF1 3109 120 - 1 GGAGCGUCUAAAACGCUCCUGCCGACCCACAGAUUGGUAUCCAGUUGAGAUGAAACAUCGCCAGCAUUACGGGGAUCUCGUCGGAAACAUUGAGAUGACAGACCAUCUUUGUAAUGGUAC (((((((.....)))))))(((((...((.((((.(((.....((((.((((...))))..))))......(..((((((..(....)..))))))..)..))))))).))...))))). ( -39.50) >DroEre_CAF1 3231 120 - 1 CGAGCGCCUAAAACGCGCAUGUCGACCCACGGAUUGGUAUCCGUCUGAGAUGAAACACCGUCAGCACUACGGGGACCUUAUAGGCCACAUUGAGAUGACCGACCAUCUAUUCGAUGGUAC ((.((((.......))))..((((.....(((((....)))))(((..((((.....((((.......))))((.((.....)))).)))).)))))))))((((((.....)))))).. ( -34.00) >consensus GGAGCGUCUAAAACGCUCCUGCCGACCCACAGAUUGGUAUCCGGUUGAGAUGAAACAUCGUCAGCAUUACGGGGAUCUCGUCGGAAACAUUGAGAUGACAGACCAUCUUUUCAAUGGUAC (((((((.....)))))))(((((......((((.(((.....(((((((((...)))).)))))....(((..((((((..(....)..))))))..))))))))))......))))). (-27.29 = -28.60 + 1.31)



| Location | 7,714,192 – 7,714,312 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7714192 120 + 22407834 AUACCAAUCUGUGGGUCGGCAGGAGCGCUUUAGACGCUCCAAAAAGGUGCCAGGACUCUGUCUCAGGUAGUAAAAUCCAUAACCAGGUAUUGCCAAAAAGGGCAACACGACAAGUAUAAU .((((....(((((((..((.(((((((....).)))))).......((((.((((...))))..))))))...)))))))....))))(((((......)))))............... ( -34.70) >DroSec_CAF1 3185 120 + 1 AUACCAAUCUGUGGGUCGGCAGGAGCGUUUUAGACGCUCCAAAAAGGUGCCAGGACUCUGUUUUAGGUAGUAAAAUGCAUAACCAGGUAUUGCCAGAAAGGGUAACACAACAAGUACAAU .((((..(((..(((((((((((((((((...)))))))).......))))..))))).......(((((((...((......))..))))))))))...))))................ ( -35.01) >DroSim_CAF1 3189 120 + 1 AUACCAAUCUGUGGGUCGGCAGGAGCGUUUUAGACGCUCCAAAAAGGUGCCAGGACUCUGUUUUAGGUAGUAAUAUCCAUAACCAGGUAUUGCCAAAAAGGGUAACACUACAAGUAUAAU .((((.(((((.(((((((((((((((((...)))))))).......))))..))))).....))))).((((((((........)))))))).......))))................ ( -33.81) >DroEre_CAF1 3311 120 + 1 AUACCAAUCCGUGGGUCGACAUGCGCGUUUUAGGCGCUCGGAAAAGGUUCCAGUGCUAUUUUUUAGGUAGUAGAAUCCAUAACCAGGUAUUGCCAGAAAGGGUAGCCCCACAAGUAUAAU ..........(((((...((..((((.......))))..((((....)))).))((((((((((.(((((((....((.......)))))))))..)))))))))))))))......... ( -33.60) >consensus AUACCAAUCUGUGGGUCGGCAGGAGCGUUUUAGACGCUCCAAAAAGGUGCCAGGACUCUGUUUUAGGUAGUAAAAUCCAUAACCAGGUAUUGCCAAAAAGGGUAACACAACAAGUAUAAU .((((....(((((((..((.(((((((.....))))))).......((((((((.....)))).))))))...)))))))....))))(((((......)))))............... (-26.92 = -27.30 + 0.38)

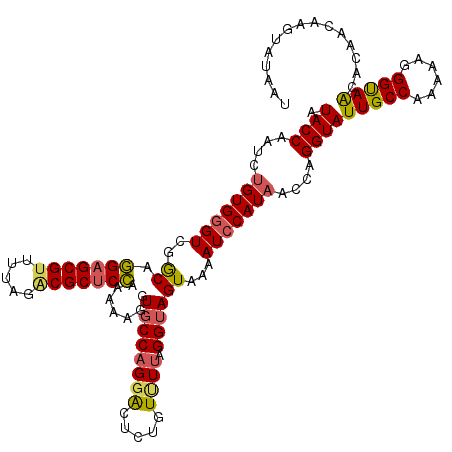
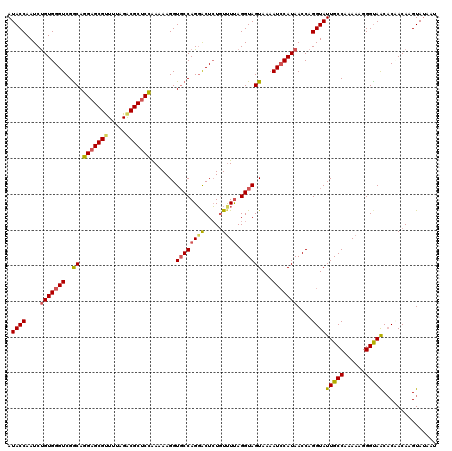
| Location | 7,714,272 – 7,714,392 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7714272 120 + 22407834 AACCAGGUAUUGCCAAAAAGGGCAACACGACAAGUAUAAUGCCUAGUACGAGGAACACUUCCUUAAAGAAGAAGUGAACUCGUAUGCACUCGAUUAUGCCCAAUAUCUAAAAGGAAAAGC ..((.((.....)).....(((((..((.....))....(((...(((((((...((((((.........))))))..))))))))))........)))))...........))...... ( -30.30) >DroSec_CAF1 3265 120 + 1 AACCAGGUAUUGCCAGAAAGGGUAACACAACAAGUACAAUGCCUAGUACGAGAAAGACUUCCUUAAAGAAGGAGUGACCUCGUAUGCACUCGAUUAUGCCCAAUAUCUAAUAGGAAAAGC ..((.((.....)).....(((((..((.....))....(((...(((((((....(((.((((....)))))))...))))))))))........)))))...........))...... ( -25.30) >DroSim_CAF1 3269 120 + 1 AACCAGGUAUUGCCAAAAAGGGUAACACUACAAGUAUAAUGCCCAAUACAAGGAACACUUCCAUAAAAAAGGAGUGACCUCGUAUGCACUCGAUUAUGCCCAAUAUCUAAUAGGAAAAGC ..((((((((((.......(((((..((.....))....))))).((((.(((..((((.((........)))))).))).))))(((........))).))))))))....))...... ( -24.80) >DroEre_CAF1 3391 120 + 1 AACCAGGUAUUGCCAGAAAGGGUAGCCCCACAAGUAUAAUCGCCAGUGCAAAGAACACUUCACGGUAGUAGAAGUGAGAAUACAUGAUUUCUUUUAUGCCGUAUACCUAGUCAGUCAAUC .((.((((((.((.((((((((....))).((.((((..((((....))......((((((((....)).)))))).)))))).)).))))).....))...)))))).))......... ( -25.30) >consensus AACCAGGUAUUGCCAAAAAGGGUAACACAACAAGUAUAAUGCCCAGUACAAGGAACACUUCCUUAAAGAAGAAGUGACCUCGUAUGCACUCGAUUAUGCCCAAUAUCUAAUAGGAAAAGC ....((((((((((......)))))........(((((((.....(((((((...((((((.........))))))..))))))).......)))))))....)))))............ (-21.15 = -21.27 + 0.13)



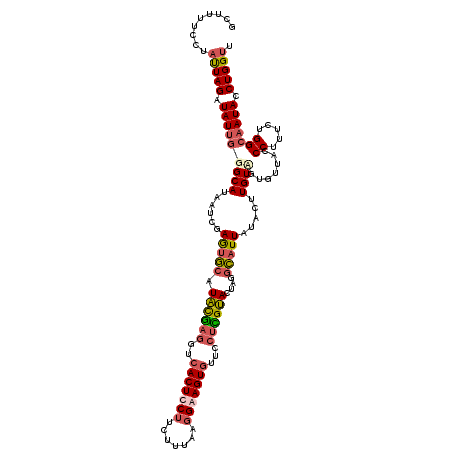
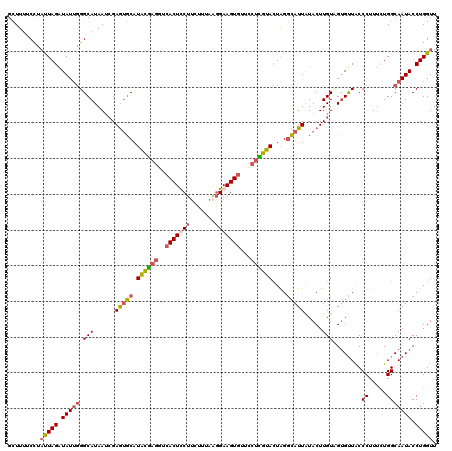
| Location | 7,714,272 – 7,714,392 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -19.16 |
| Energy contribution | -20.60 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7714272 120 - 22407834 GCUUUUCCUUUUAGAUAUUGGGCAUAAUCGAGUGCAUACGAGUUCACUUCUUCUUUAAGGAAGUGUUCCUCGUACUAGGCAUUAUACUUGUCGUGUUGCCCUUUUUGGCAAUACCUGGUU ..........((((......((((......(((((.((((((..((((((((.....))))))))...))))))....))))).....))))((((((((......)))))))))))).. ( -37.00) >DroSec_CAF1 3265 120 - 1 GCUUUUCCUAUUAGAUAUUGGGCAUAAUCGAGUGCAUACGAGGUCACUCCUUCUUUAAGGAAGUCUUUCUCGUACUAGGCAUUGUACUUGUUGUGUUACCCUUUCUGGCAAUACCUGGUU .........(((((.((((((((((((.(((((((((((((((..(((((((....)))).)))...)))))))........)))))))))))))))..((.....))))))).))))). ( -33.20) >DroSim_CAF1 3269 120 - 1 GCUUUUCCUAUUAGAUAUUGGGCAUAAUCGAGUGCAUACGAGGUCACUCCUUUUUUAUGGAAGUGUUCCUUGUAUUGGGCAUUAUACUUGUAGUGUUACCCUUUUUGGCAAUACCUGGUU .........(((((.(((((.((((......))))((((((((.((((((........)).))))..)))))))).(((((((((....))))))...))).......))))).))))). ( -31.70) >DroEre_CAF1 3391 120 - 1 GAUUGACUGACUAGGUAUACGGCAUAAAAGAAAUCAUGUAUUCUCACUUCUACUACCGUGAAGUGUUCUUUGCACUGGCGAUUAUACUUGUGGGGCUACCCUUUCUGGCAAUACCUGGUU ........((((((((((...((.....(((((....(((..(((((.........(((..(((((.....))))).))).........)))))..)))..))))).)).)))))))))) ( -33.07) >consensus GCUUUUCCUAUUAGAUAUUGGGCAUAAUCGAGUGCAUACGAGGUCACUCCUUCUUUAAGGAAGUGUUCCUCGUACUAGGCAUUAUACUUGUAGUGUUACCCUUUCUGGCAAUACCUGGUU .........(((((.(((((((((......(((((.((((((..(((((((.......)))))))...))))))....))))).....)))).......((.....))))))).))))). (-19.16 = -20.60 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:04 2006