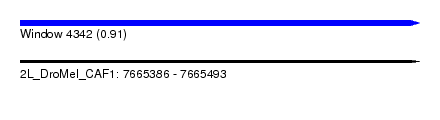
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,665,386 – 7,665,493 |
| Length | 107 |
| Max. P | 0.910972 |

| Location | 7,665,386 – 7,665,493 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -14.24 |
| Energy contribution | -16.18 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
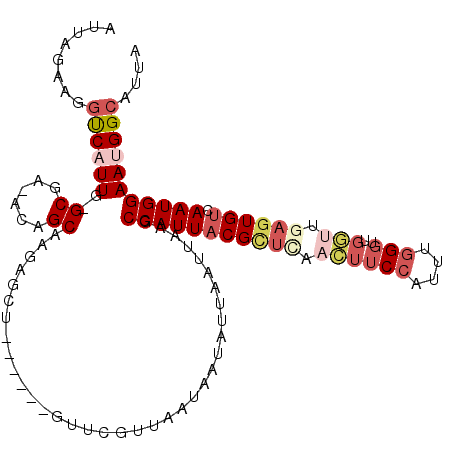
>2L_DroMel_CAF1 7665386 107 + 22407834 AUUAGAAGGUCAUUG-GCGA-ACAGCAAGAGCU------GUUCGUUAAUAAUAUUAAUUAAGCCAUUACGCUCAACUUCCAUUUGGGUUGGUUGAGUGUCAAUGGAAUGGCAUUA ........(((((((-((((-(((((....)))------)))))))))).............((((((((((((((((((....)))..)))))))))).)))))...))).... ( -42.00) >DroSec_CAF1 10572 107 + 1 AUUAGAAGGUCAUUG-GCGA-ACAGCAAGUGCU------GUUCGUUAAUAAUAUUAAUUAAGCCAUUACGCUCAACUUCCAUUUGGGUUGGUUGAGUGUCAAUGGAAUGGCAUUA ........(((((((-((((-(((((....)))------)))))))))).............((((((((((((((((((....)))..)))))))))).)))))...))).... ( -42.30) >DroEre_CAF1 18780 113 + 1 AUUAGAAGGUCAUUG-GCGA-ACAGCAAGAGCUGUUGCUGUUCGUUAAUAAUAUUAAUUAAGCCAUUACGCUCUACUUCCAUUUGGGUUGGUUGAGUGUCAAUGGAAUGGAAUUA ...........((((-((((-(((((((......))))))))))))))).....(((((...(((((((((((.((((((....)))..))).)))))).))))).....))))) ( -37.80) >DroYak_CAF1 10832 107 + 1 AUUAGAAGGUCAUUG-GCGA-ACAGCAAGAGCU------GUUCGGUAAUAAUAUUAAUUAAGCCAUUACGCUUCACUUCCAUUUGGGUUCGUUGAGUGUCAAUGGAAUGGCAUUA ........((((((.-.(((-(((((....)))------)))))..................(((((((((((.((.(((....)))...)).)))))).))))))))))).... ( -33.20) >DroAna_CAF1 14066 108 + 1 AUUAGAAGGUCAUUGGGCGA-ACAGCUAGAACG------CUCGGUUAAUAAUAUUAAUUAAGCCAUUACGUUCUAGUUCCAUUUUGUUGGCCUAAGUGUCAAUGGAAAGGCAUUA .............(((((.(-((((((((((((------((..((((((...))))))..)))......)))))))).......)))).)))))((((((........)))))). ( -29.91) >DroPer_CAF1 11527 88 + 1 AUUAGUAGGCCAUUG-GCGAAACAGCCAAAGCG------UUUGGUUAAUAACAUUAAUUAAGCCAUUUCG--------------------AGUGGCUGUCAAUGGAAAGGCAUUA ....((...((((((-((.....((((((....------.))))))..............((((((....--------------------.))))))))))))))....)).... ( -26.20) >consensus AUUAGAAGGUCAUUG_GCGA_ACAGCAAGAGCU______GUUCGUUAAUAAUAUUAAUUAAGCCAUUACGCUCAACUUCCAUUUGGGUUGGUUGAGUGUCAAUGGAAUGGCAUUA ........((((((..((......))....................................(((((((((((.((((((....)))..))).)))))).))))))))))).... (-14.24 = -16.18 + 1.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:49 2006