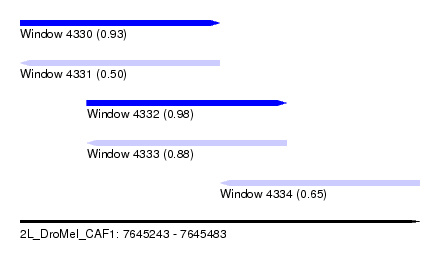
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,645,243 – 7,645,483 |
| Length | 240 |
| Max. P | 0.977181 |

| Location | 7,645,243 – 7,645,363 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -57.38 |
| Consensus MFE | -43.05 |
| Energy contribution | -44.42 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7645243 120 + 22407834 GGAAGUUGUUUGGGGAACUCUCGCGACAGGGCGAUGAAAUCGCCGAGUCCUGGGAGACCAUACUCCUAGGAAUGGUGGCUAGUUCCAGGCAGGUUUCAUUACCACCUGUGGUGGUGGCUA ((((.(((((...((((((...((.((..((((((...))))))...(((((((((......)))))))))...)).)).)))))).))))).)))).((((((((...))))))))... ( -52.60) >DroPse_CAF1 185234 120 + 1 GGAAGUUGCUCGGCGAACUCUCGCGGCAGGGCGAGGAGAUGGCCGAGUCCUGGGAGACCAUGCUCCUGGGAACGGUGGCCAGCUCCAGGCAGGUCUCGUUGCCGCCCGUCGUCGUGGCUA ...(((..(...((((....))))(((.((((..((((.((((((..(((..((((......))))..)))....)))))).)))).(((((......))))))))))))...)..))). ( -62.40) >DroAna_CAF1 156851 120 + 1 GGAAGUUGUUGGGGGAACUCUCGCGACAUGGCGACGAUAUGGCCGAGUCCUGCGAUACCAUACUCCUGGGUAUGGUGGCCAACUCCAGGCAGGUCUCGUUGCCACCCGUUGUGGUGGCUA ...((((((((.(((....))).))))((.((((((...((((((((.(((((...((((((((....))))))))((......))..))))).))))..))))..)))))).)))))). ( -51.40) >DroPer_CAF1 182056 120 + 1 GGAAGUUGCUCGGCGAGCUCUCGCGGCAGGGCGAGGAGAUGGCCGAGUCCUGGGAGACCAUGCUCCUGGGAACGGUGGCCAGCUCCAGGCAGGUCUCGUUGCCGCCCGUCGUCGUGGCUA ...(((..(...((((....))))(((.((((..((((.((((((..(((..((((......))))..)))....)))))).)))).(((((......))))))))))))...)..))). ( -63.10) >consensus GGAAGUUGCUCGGCGAACUCUCGCGACAGGGCGAGGAGAUGGCCGAGUCCUGGGAGACCAUACUCCUGGGAACGGUGGCCAGCUCCAGGCAGGUCUCGUUGCCACCCGUCGUCGUGGCUA (..(.(((((.((((..(((((((......)))).))).((((((.((((((((((......)))))))).))..)))))).)))).))))).)..)...((((((......)))))).. (-43.05 = -44.42 + 1.38)

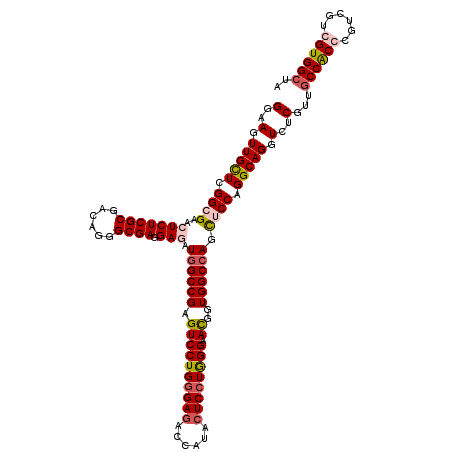
| Location | 7,645,243 – 7,645,363 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -32.91 |
| Energy contribution | -34.60 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7645243 120 - 22407834 UAGCCACCACCACAGGUGGUAAUGAAACCUGCCUGGAACUAGCCACCAUUCCUAGGAGUAUGGUCUCCCAGGACUCGGCGAUUUCAUCGCCCUGUCGCGAGAGUUCCCCAAACAACUUCC ..((((((......))))))..............((((((.((.((...((((.((((......)))).))))...((((((...))))))..)).))...))))))............. ( -39.50) >DroPse_CAF1 185234 120 - 1 UAGCCACGACGACGGGCGGCAACGAGACCUGCCUGGAGCUGGCCACCGUUCCCAGGAGCAUGGUCUCCCAGGACUCGGCCAUCUCCUCGCCCUGCCGCGAGAGUUCGCCGAGCAACUUCC ..(((.((....)))))(....)((((((((((((((((.(....).)))))..)).))).))))))...(((((((((...(((.((((......)))))))...)))))).....))) ( -48.30) >DroAna_CAF1 156851 120 - 1 UAGCCACCACAACGGGUGGCAACGAGACCUGCCUGGAGUUGGCCACCAUACCCAGGAGUAUGGUAUCGCAGGACUCGGCCAUAUCGUCGCCAUGUCGCGAGAGUUCCCCCAACAACUUCC ..((((((......))))))..((((.(((((..((......))(((((((......)))))))...))))).)))).........((((......))))(((((........))))).. ( -41.80) >DroPer_CAF1 182056 120 - 1 UAGCCACGACGACGGGCGGCAACGAGACCUGCCUGGAGCUGGCCACCGUUCCCAGGAGCAUGGUCUCCCAGGACUCGGCCAUCUCCUCGCCCUGCCGCGAGAGCUCGCCGAGCAACUUCC ..((..((.(((.((((((((........)))).((((.(((((...((((....))))..(((((....))))).))))).))))..))))....((....))))).)).))....... ( -48.50) >consensus UAGCCACCACGACGGGCGGCAACGAGACCUGCCUGGAGCUGGCCACCAUUCCCAGGAGCAUGGUCUCCCAGGACUCGGCCAUCUCCUCGCCCUGCCGCGAGAGUUCCCCGAACAACUUCC ..(((((........)))))...(((.((((...((((...((((..((((....)))).)))))))))))).)))(((...(((.((((......)))))))...)))........... (-32.91 = -34.60 + 1.69)


| Location | 7,645,283 – 7,645,403 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -57.12 |
| Consensus MFE | -46.21 |
| Energy contribution | -46.15 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7645283 120 + 22407834 CGCCGAGUCCUGGGAGACCAUACUCCUAGGAAUGGUGGCUAGUUCCAGGCAGGUUUCAUUACCACCUGUGGUGGUGGCUAUGAUGACAUCCUCGACGCCCUCGUCAAUGGGCGAUGAAAU .((((..(((((((((......)))))))))....))))....((.(((...(((((((..(((((......)))))..)))).)))..))).))(((((........)))))....... ( -50.30) >DroPse_CAF1 185274 120 + 1 GGCCGAGUCCUGGGAGACCAUGCUCCUGGGAACGGUGGCCAGCUCCAGGCAGGUCUCGUUGCCGCCCGUCGUCGUGGCUAUGAUGAUGUCCUCGACGCCCUCGUCGAUGGGCGACGAGAU (((((..(((..((((......))))..)))....))))).((.....))..((((((((((((..(((((((((....)))))))))..)((((((....))))))..))))))))))) ( -61.50) >DroAna_CAF1 156891 120 + 1 GGCCGAGUCCUGCGAUACCAUACUCCUGGGUAUGGUGGCCAACUCCAGGCAGGUCUCGUUGCCACCCGUUGUGGUGGCUAUGAUGAUGUCCUCGACACCUUCGUCGAUGGGCGAGGAGAU ((((..(((....)))((((((((....))))))))))))..((((......(((((((.((((((......)))))).)))).)))(((((((((......))))).))))..)))).. ( -55.20) >DroPer_CAF1 182096 120 + 1 GGCCGAGUCCUGGGAGACCAUGCUCCUGGGAACGGUGGCCAGCUCCAGGCAGGUCUCGUUGCCGCCCGUCGUCGUGGCUAUGAUGAUGUCCUCGACGCCCUCGUCGAUGGGCGACGAGAU (((((..(((..((((......))))..)))....))))).((.....))..((((((((((((..(((((((((....)))))))))..)((((((....))))))..))))))))))) ( -61.50) >consensus GGCCGAGUCCUGGGAGACCAUACUCCUGGGAACGGUGGCCAGCUCCAGGCAGGUCUCGUUGCCACCCGUCGUCGUGGCUAUGAUGAUGUCCUCGACGCCCUCGUCGAUGGGCGACGAGAU (((((.((((((((((......)))))))).))..)))))..(((.(((.(.(((((((.((((((......)))))).)))).))).))))...(((((........)))))..))).. (-46.21 = -46.15 + -0.06)

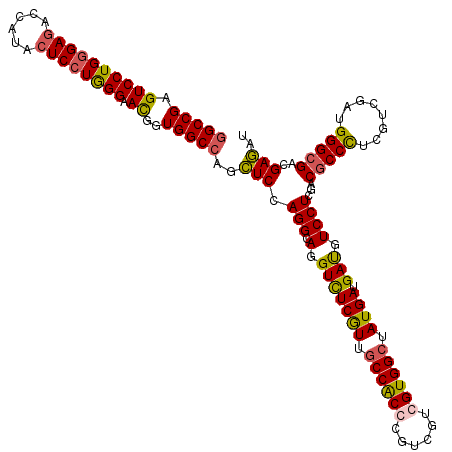

| Location | 7,645,283 – 7,645,403 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -49.33 |
| Consensus MFE | -39.33 |
| Energy contribution | -40.45 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7645283 120 - 22407834 AUUUCAUCGCCCAUUGACGAGGGCGUCGAGGAUGUCAUCAUAGCCACCACCACAGGUGGUAAUGAAACCUGCCUGGAACUAGCCACCAUUCCUAGGAGUAUGGUCUCCCAGGACUCGGCG ........((((........))))((((((.......((((.((((((......)))))).))))......(((((.....((((..((((....)))).))))...))))).)))))). ( -46.50) >DroPse_CAF1 185274 120 - 1 AUCUCGUCGCCCAUCGACGAGGGCGUCGAGGACAUCAUCAUAGCCACGACGACGGGCGGCAACGAGACCUGCCUGGAGCUGGCCACCGUUCCCAGGAGCAUGGUCUCCCAGGACUCGGCC .(((((.(((((........))))).)))))...........(((.((....)))))(((..((((.((((...((((...((((..((((....)))).)))))))))))).))))))) ( -50.90) >DroAna_CAF1 156891 120 - 1 AUCUCCUCGCCCAUCGACGAAGGUGUCGAGGACAUCAUCAUAGCCACCACAACGGGUGGCAACGAGACCUGCCUGGAGUUGGCCACCAUACCCAGGAGUAUGGUAUCGCAGGACUCGGCC ...((((((..((((......)))).))))))..........((((((......))))))..((((.(((((..((......))(((((((......)))))))...))))).))))... ( -49.00) >DroPer_CAF1 182096 120 - 1 AUCUCGUCGCCCAUCGACGAGGGCGUCGAGGACAUCAUCAUAGCCACGACGACGGGCGGCAACGAGACCUGCCUGGAGCUGGCCACCGUUCCCAGGAGCAUGGUCUCCCAGGACUCGGCC .(((((.(((((........))))).)))))...........(((.((....)))))(((..((((.((((...((((...((((..((((....)))).)))))))))))).))))))) ( -50.90) >consensus AUCUCGUCGCCCAUCGACGAGGGCGUCGAGGACAUCAUCAUAGCCACCACGACGGGCGGCAACGAGACCUGCCUGGAGCUGGCCACCAUUCCCAGGAGCAUGGUCUCCCAGGACUCGGCC .((((((((.....))))))))..((((((..(.........(((((........)))))...((((((..((((((((........))))).))).....))))))...)..)))))). (-39.33 = -40.45 + 1.12)



| Location | 7,645,363 – 7,645,483 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -33.44 |
| Energy contribution | -32.75 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7645363 120 - 22407834 AGGACUCAGCCUUUGGCUCUCUGACUGACAACGACCUGUCCAUUGGAUCGUCCAGCAUUAGGUUGAGCAGCUUUCAGUCCAUUUCAUCGCCCAUUGACGAGGGCGUCGAGGAUGUCAUCA .(((((.((((...))))......(((....(((((((....((((.....))))...)))))))..))).....))))).((((..(((((........)))))..))))......... ( -36.20) >DroPse_CAF1 185354 117 - 1 AGGACUCGGCCUUCGGCUCGCUCACCGACAACGACCUGUCCAUCG---CGUCGAGCAUCCGGCUGAGCAGCUUCCAGUCCAUCUCGUCGCCCAUCGACGAGGGCGUCGAGGACAUCAUCA .(((((.((.....((((.(((((((((((......))))....(---(.....))....)).))))))))).))))))).(((((.(((((........))))).)))))......... ( -46.70) >DroAna_CAF1 156971 120 - 1 AGGACUCGGCCUUCGGUUCUCUGACCGACAAUGACCUCUCCAUUGGUUCGUCCAGCAUCCGGCUGAGCAGUUUCCAGUCCAUCUCCUCGCCCAUCGACGAAGGUGUCGAGGACAUCAUCA .(((((.((...((((((....))))))(((((.......)))))....(..((((.....))))..).....)))))))...((((((..((((......)))).))))))........ ( -38.50) >DroPer_CAF1 182176 117 - 1 AGGACUCGGCCUUCGGCUCGCUCACCGACAACGACCUGUCCAUCG---CGUCGAGCAUCCGGCUGAGCAGCUUCCAGUCCAUCUCGUCGCCCAUCGACGAGGGCGUCGAGGACAUCAUCA .(((((.((.....((((.(((((((((((......))))....(---(.....))....)).))))))))).))))))).(((((.(((((........))))).)))))......... ( -46.70) >consensus AGGACUCGGCCUUCGGCUCGCUCACCGACAACGACCUGUCCAUCG___CGUCCAGCAUCCGGCUGAGCAGCUUCCAGUCCAUCUCGUCGCCCAUCGACGAGGGCGUCGAGGACAUCAUCA .(((((.(((.(((((((.((((..(((...(((........)))..)))..))))....)))))))..)))...))))).(((((.(((((........))))).)))))......... (-33.44 = -32.75 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:41 2006