
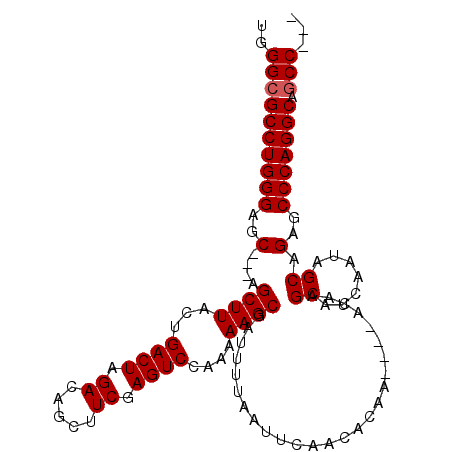
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,626,623 – 7,626,728 |
| Length | 105 |
| Max. P | 0.974004 |

| Location | 7,626,623 – 7,626,728 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7626623 105 + 22407834 UGGGCGCCUGGGAGCAGCAGCUUACUGACUAGACAGCUUCGAGUCCAAAAAGCGAUUUUAAUUCAACACAAACAAACAAGCAUCAAUAGCAGAGCCCAGGCAGCC--- ..((((((((((..(.((.((((...((((.((.....)).))))....))))((.......))........................)).)..))))))).)))--- ( -30.80) >DroSec_CAF1 126373 98 + 1 UGGGCGCCUGGGAGC---AGCUUACUGACUAGACAGCUUCGAGUCCAAAAAGCGAUUUUAAUUCAACACAA----ACAAGCAUCAAUAGCAGAGCCCAGGCAGCC--- ..((((((((((..(---.((((...((((.((.....)).))))....))))..................----....((.......)).)..))))))).)))--- ( -29.80) >DroSim_CAF1 132279 98 + 1 UGGGCGCCUGGGAGC---AGCUUACUGACUAGACAGCUUCGAGUCCAAAAAGCGAUUUUAAUUCAACACAA----ACAAGCAUCAAUAGCAGAGCCCAGGCAGCC--- ..((((((((((..(---.((((...((((.((.....)).))))....))))..................----....((.......)).)..))))))).)))--- ( -29.80) >DroYak_CAF1 129699 101 + 1 UGGGCGCCUGGGAGC---AGCUUACUGACUAGACAGCUUCGAGUCCAAAAAGCGAUUUUAAUUCAACACAA----ACAGGCAUCAAUAGCAGAGCCCAGGCAACCCAG ((((.(((((((..(---.((((...((((.((.....)).))))....))))..................----....((.......)).)..)))))))..)))). ( -31.00) >consensus UGGGCGCCUGGGAGC___AGCUUACUGACUAGACAGCUUCGAGUCCAAAAAGCGAUUUUAAUUCAACACAA____ACAAGCAUCAAUAGCAGAGCCCAGGCAGCC___ ..((((((((((..(....((((...((((.((.....)).))))....))))..........................((.......)).)..))))))).)))... (-26.98 = -27.23 + 0.25)


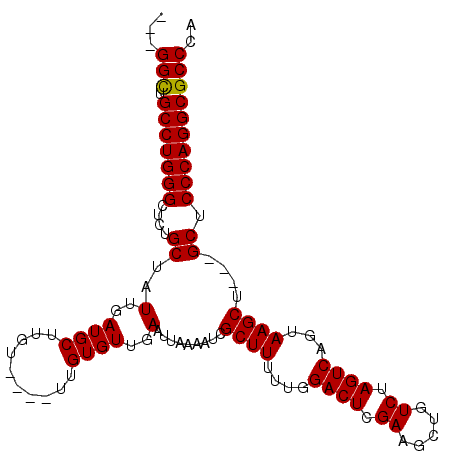
| Location | 7,626,623 – 7,626,728 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -32.01 |
| Energy contribution | -31.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7626623 105 - 22407834 ---GGCUGCCUGGGCUCUGCUAUUGAUGCUUGUUUGUUUGUGUUGAAUUAAAAUCGCUUUUUGGACUCGAAGCUGUCUAGUCAGUAAGCUGCUGCUCCCAGGCGCCCA ---(((.(((((((..(.((..(..((((..........))))..).........((((....((((.((.....)).))))...)))).)).)..)))))))))).. ( -33.70) >DroSec_CAF1 126373 98 - 1 ---GGCUGCCUGGGCUCUGCUAUUGAUGCUUGU----UUGUGUUGAAUUAAAAUCGCUUUUUGGACUCGAAGCUGUCUAGUCAGUAAGCU---GCUCCCAGGCGCCCA ---(((.(((((((..(.(((.(..((((....----..))))..).........(((..((((((........))))))..))).))).---)..)))))))))).. ( -35.30) >DroSim_CAF1 132279 98 - 1 ---GGCUGCCUGGGCUCUGCUAUUGAUGCUUGU----UUGUGUUGAAUUAAAAUCGCUUUUUGGACUCGAAGCUGUCUAGUCAGUAAGCU---GCUCCCAGGCGCCCA ---(((.(((((((..(.(((.(..((((....----..))))..).........(((..((((((........))))))..))).))).---)..)))))))))).. ( -35.30) >DroYak_CAF1 129699 101 - 1 CUGGGUUGCCUGGGCUCUGCUAUUGAUGCCUGU----UUGUGUUGAAUUAAAAUCGCUUUUUGGACUCGAAGCUGUCUAGUCAGUAAGCU---GCUCCCAGGCGCCCA ..((((.(((((((..(.(((((((((((...(----(((........))))...))....(((((........))))))))))).))).---)..))))))))))). ( -37.10) >consensus ___GGCUGCCUGGGCUCUGCUAUUGAUGCUUGU____UUGUGUUGAAUUAAAAUCGCUUUUUGGACUCGAAGCUGUCUAGUCAGUAAGCU___GCUCCCAGGCGCCCA ...(((.(((((((....((..(..((((..........))))..).........((((....((((.((.....)).))))...))))....)).)))))))))).. (-32.01 = -31.82 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:32 2006