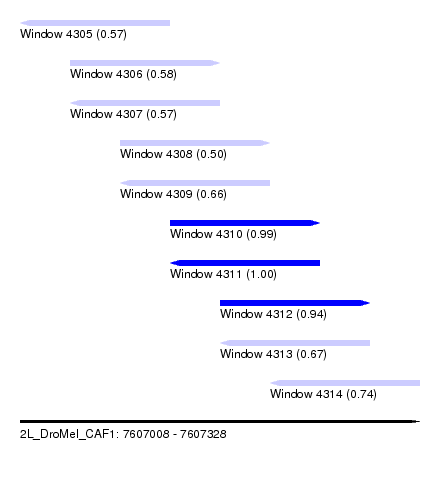
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,607,008 – 7,607,328 |
| Length | 320 |
| Max. P | 0.998865 |

| Location | 7,607,008 – 7,607,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -32.66 |
| Energy contribution | -33.14 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607008 120 - 22407834 UAUCUUCCGAGAAUUGGAACUGGUGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAACUUGGUUUGGGUCACUGAAGCGUCGGGAUCUGGUCCCAGA ....((((((...(((((.(((((((((((....)))))(((.....)))))).))))))))...(((((((((((((......).)))))..)))))))..))))))((((....)))) ( -38.50) >DroSec_CAF1 106639 120 - 1 UAUCUUCCGAGGAUUGGAACUGGUGGAUGCAAGCGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAACUUGGUUUGGGUCACUGAGGCGUCGGGAUCCGGUCCCAGA ........(.((((((((.(((((((((((....)))))(((.....)))))).)))(((.((..(((((((((((((......).)))))..)))))))..)).))))))))))))... ( -41.00) >DroSim_CAF1 112812 120 - 1 UAUCUUCCGAGGAUUGGAACUGGUGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAACUUGGUUUGGGUCACUGAAGCGUCGGGAUCCGGUCCCAGA ........(.((((((((.(((((((((((....)))))(((.....)))))).)))(((.((..(((((((((((((......).)))))..)))))))..)).))))))))))))... ( -41.30) >DroEre_CAF1 106940 120 - 1 UACCUUCCGAGUAUAGGAACUGGUGGAUACAAGCGCAUCGUCUCCUUGAUCACGCAGUCGAGAUACUGAAGUUCCAGCAACUUGGAUUGGGUCACCGAAGCGUCGGGAUCCGGUCCCAGA ....((((.......)))).((.((((.((..(((.((((......))))..)))(((......)))...)))))).)).((.((((((((((.((((....)))))))))))))).)). ( -38.10) >DroYak_CAF1 109511 120 - 1 UACCUACCGAGAAUGGGAACUGGAGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCGAGAUACUUGAGUUCCAGCAACUUGGUUUGGGUCACUGGAGCGUCGGGAUCCGUUCCUAGA .((((((((((....((((((.((((((((....))))).....((((((......))))))...))).)))))).....))))))..))))..((((((((........)))))).)). ( -44.00) >consensus UAUCUUCCGAGAAUUGGAACUGGUGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAACUUGGUUUGGGUCACUGAAGCGUCGGGAUCCGGUCCCAGA ......(((((..((((((((((.((((((....)))))((((..(((......)))...)))).).))))))))))...)))))..((((((.((((....))))))))))........ (-32.66 = -33.14 + 0.48)

| Location | 7,607,048 – 7,607,168 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -27.26 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607048 120 + 22407834 UUGUUGGAACUAAAGUAUCUGGACUGCGUGAUCAAGGAGACGAUGCGUUUGCAUCCACCAGUUCCAAUUCUCGGAAGAUACAUUCCCGAGGACUUGAAAAUUGGCGAAAUAACCAUUCCU ((((..(.....((((...(((((((.(((.....(....)(((((....))))))))))).))))..((((((((......)).)))))))))).....)..))))............. ( -32.90) >DroSec_CAF1 106679 120 + 1 UUGUUGGAACUAAAGUAUCUGGACUGCGUGAUCAAGGAGACGAUGCGCUUGCAUCCACCAGUUCCAAUCCUCGGAAGAUACAUUCCCGAGGACUUGCAAAUUUGCGAAAAAACCAUUCCU .....((((..........(((((((.(((.....(....)(((((....))))))))))).)))).(((((((((......)).))))))).(((((....)))))........)))). ( -36.50) >DroSim_CAF1 112852 120 + 1 UUGUUGGAACUAAAGUAUCUGGACUGCGUGAUCAAGGAGACGAUGCGUUUGCAUCCACCAGUUCCAAUCCUCGGAAGAUACAUUCCCGAGGACCUGCAAAUUGGGGAAAAAACCAUUCCU ((((.((............(((((((.(((.....(....)(((((....))))))))))).)))).(((((((((......)).))))))))).))))....(((((.......))))) ( -38.10) >DroEre_CAF1 106980 120 + 1 UUGCUGGAACUUCAGUAUCUCGACUGCGUGAUCAAGGAGACGAUGCGCUUGUAUCCACCAGUUCCUAUACUCGGAAGGUACAUUCCCGAGGACCUGAACAUUGGCGAUAAAACCAUUCCC ((((..(...(((((..(((((...((((.(((..(....)))))))).((((((..(((((......))).))..))))))....)))))..)))))..)..))))............. ( -33.20) >DroYak_CAF1 109551 120 + 1 UUGCUGGAACUCAAGUAUCUCGACUGCGUGAUCAAGGAGACGAUGCGUUUGCAUCCUCCAGUUCCCAUUCUCGGUAGGUACAUUCCCGAGGACUUGCACAUCGGCGAUAAGACCAUUCCC (((((((((((...(.(((.((....)).))))..((((..(((((....)))))))))))))))..(((((((.((.....)).)))))))..........)))))............. ( -33.10) >consensus UUGUUGGAACUAAAGUAUCUGGACUGCGUGAUCAAGGAGACGAUGCGUUUGCAUCCACCAGUUCCAAUCCUCGGAAGAUACAUUCCCGAGGACUUGCAAAUUGGCGAAAAAACCAUUCCU .....((((...........((((((.(((.....(....)(((((....))))))))))))))...(((((((..((.....))))))))).......................)))). (-27.26 = -27.50 + 0.24)

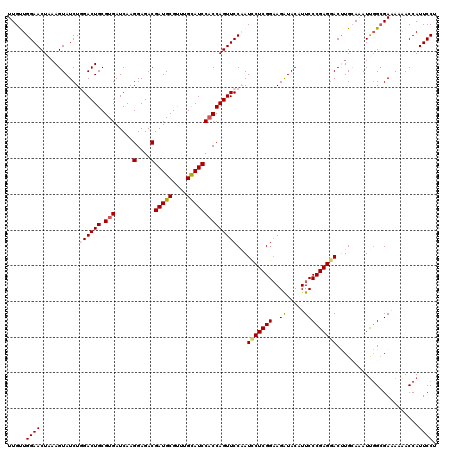
| Location | 7,607,048 – 7,607,168 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -27.60 |
| Energy contribution | -29.36 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607048 120 - 22407834 AGGAAUGGUUAUUUCGCCAAUUUUCAAGUCCUCGGGAAUGUAUCUUCCGAGAAUUGGAACUGGUGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAA .((((((((......))).......((((.(((((((.......)))))))..(((((.(((((((((((....)))))(((.....)))))).))))))))..))))..)))))..... ( -36.20) >DroSec_CAF1 106679 120 - 1 AGGAAUGGUUUUUUCGCAAAUUUGCAAGUCCUCGGGAAUGUAUCUUCCGAGGAUUGGAACUGGUGGAUGCAAGCGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAA .(((((.......(((((....))).(((((((((((.......)))))))))))(((.(((((((((((....)))))(((.....)))))).)))))).)).......)))))..... ( -42.04) >DroSim_CAF1 112852 120 - 1 AGGAAUGGUUUUUUCCCCAAUUUGCAGGUCCUCGGGAAUGUAUCUUCCGAGGAUUGGAACUGGUGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAA .(((((.......((...........(((((((((((.......)))))))))))(((.(((((((((((....)))))(((.....)))))).)))))).)).......)))))..... ( -39.24) >DroEre_CAF1 106980 120 - 1 GGGAAUGGUUUUAUCGCCAAUGUUCAGGUCCUCGGGAAUGUACCUUCCGAGUAUAGGAACUGGUGGAUACAAGCGCAUCGUCUCCUUGAUCACGCAGUCGAGAUACUGAAGUUCCAGCAA .(((((..(..((((.(((..((((..((.(((((((.......))))))).))..))))...)))......(((.((((......))))..)))......))))..)..)))))..... ( -33.90) >DroYak_CAF1 109551 120 - 1 GGGAAUGGUCUUAUCGCCGAUGUGCAAGUCCUCGGGAAUGUACCUACCGAGAAUGGGAACUGGAGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCGAGAUACUUGAGUUCCAGCAA .....((((......))))...(((.....(((((...........)))))....((((((.((((((((....))))).....((((((......))))))...))).)))))).))). ( -38.20) >consensus AGGAAUGGUUUUUUCGCCAAUUUGCAAGUCCUCGGGAAUGUAUCUUCCGAGAAUUGGAACUGGUGGAUGCAAACGCAUCGUCUCCUUGAUCACGCAGUCCAGAUACUUUAGUUCCAACAA .((((((((......))).......((((.(((((((.......)))))))..(((((.(((((((((((....)))))(((.....)))))).))))))))..))))..)))))..... (-27.60 = -29.36 + 1.76)

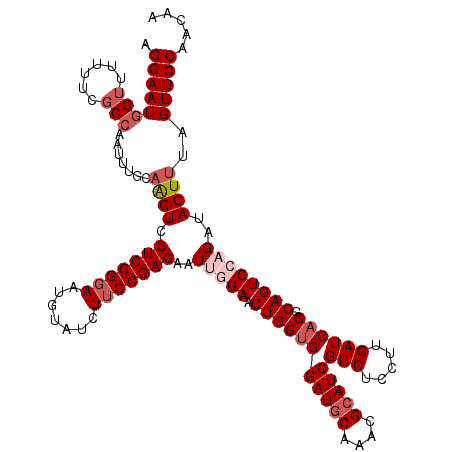
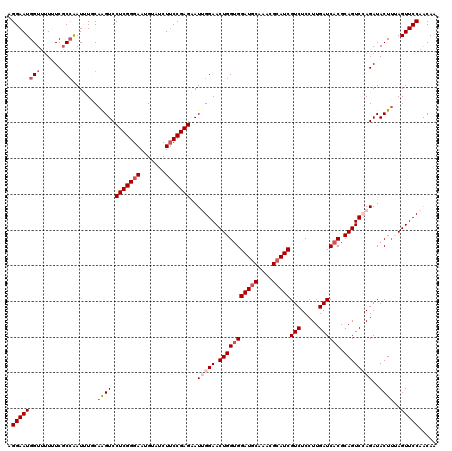
| Location | 7,607,088 – 7,607,208 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -27.01 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607088 120 + 22407834 CGAUGCGUUUGCAUCCACCAGUUCCAAUUCUCGGAAGAUACAUUCCCGAGGACUUGAAAAUUGGCGAAAUAACCAUUCCUGGCAACACAAGCAUAUUACUAAUGCCCUACUAUGUCUACC .(((((....)))))(.((((((.((((((((((((......)).))))))).)))..)))))).)..............((((......((((.......)))).......)))).... ( -25.62) >DroSec_CAF1 106719 120 + 1 CGAUGCGCUUGCAUCCACCAGUUCCAAUCCUCGGAAGAUACAUUCCCGAGGACUUGCAAAUUUGCGAAAAAACCAUUCCUGGCAACACAAGCAUUUUACUAAUGCCGUACUAUGUCUACC .(((((....))))).....((((((.(((((((((......)).))))))).(((((....)))))............))).)))....(((((.....)))))............... ( -28.80) >DroSim_CAF1 112892 120 + 1 CGAUGCGUUUGCAUCCACCAGUUCCAAUCCUCGGAAGAUACAUUCCCGAGGACCUGCAAAUUGGGGAAAAAACCAUUCCUGGCAACACAAGCAUUUUACUAAUGCCGUACUAUGUCUACC .((((.((((...(((.((((((.((.(((((((((......)).)))))))..))..)))))))))..))))))))...((((......(((((.....))))).......)))).... ( -31.82) >DroEre_CAF1 107020 120 + 1 CGAUGCGCUUGUAUCCACCAGUUCCUAUACUCGGAAGGUACAUUCCCGAGGACCUGAACAUUGGCGAUAAAACCAUUCCCGGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACC .(((((....)))))..(((((((.....(((((((......)).))))).....))))..))).((((...........(....)....(((((.....))))).......)))).... ( -23.40) >DroYak_CAF1 109591 120 + 1 CGAUGCGUUUGCAUCCUCCAGUUCCCAUUCUCGGUAGGUACAUUCCCGAGGACUUGCACAUCGGCGAUAAGACCAUUCCCGGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACC .(((((....))))).................(((((((....(((((((........).)))).))....)))......((((......(((((.....))))).......)))))))) ( -25.42) >consensus CGAUGCGUUUGCAUCCACCAGUUCCAAUCCUCGGAAGAUACAUUCCCGAGGACUUGCAAAUUGGCGAAAAAACCAUUCCUGGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACC .(((((....)))))(.((((((....(((((((..((.....)))))))))......)))))).)..............((((......(((((.....))))).......)))).... (-21.60 = -22.24 + 0.64)

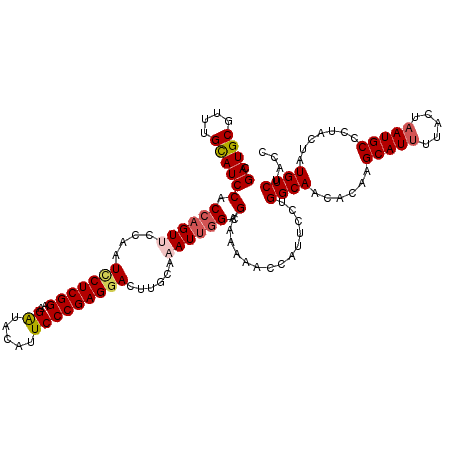
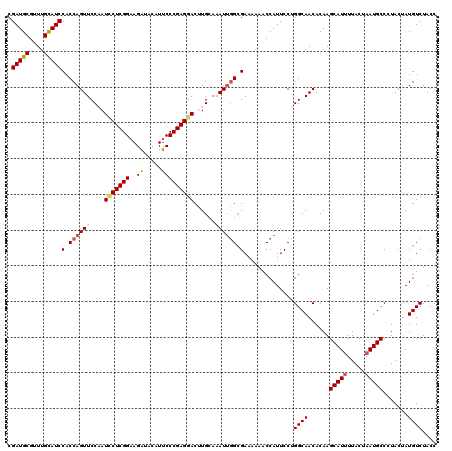
| Location | 7,607,088 – 7,607,208 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -29.34 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607088 120 - 22407834 GGUAGACAUAGUAGGGCAUUAGUAAUAUGCUUGUGUUGCCAGGAAUGGUUAUUUCGCCAAUUUUCAAGUCCUCGGGAAUGUAUCUUCCGAGAAUUGGAACUGGUGGAUGCAAACGCAUCG ....((((((....(((((.......)))))))))))(((((...((((......))))..((((((...(((((((.......)))))))..))))))))))).(((((....))))). ( -37.50) >DroSec_CAF1 106719 120 - 1 GGUAGACAUAGUACGGCAUUAGUAAAAUGCUUGUGUUGCCAGGAAUGGUUUUUUCGCAAAUUUGCAAGUCCUCGGGAAUGUAUCUUCCGAGGAUUGGAACUGGUGGAUGCAAGCGCAUCG ....((((((....((((((.....))))))))))))(((((.............(((....))).(((((((((((.......)))))))))))....))))).(((((....))))). ( -40.60) >DroSim_CAF1 112892 120 - 1 GGUAGACAUAGUACGGCAUUAGUAAAAUGCUUGUGUUGCCAGGAAUGGUUUUUUCCCCAAUUUGCAGGUCCUCGGGAAUGUAUCUUCCGAGGAUUGGAACUGGUGGAUGCAAACGCAUCG (((((.((((....((((((.....))))))))))))))).((((.......))))(((.(((...(((((((((((.......))))))))))).))).)))..(((((....))))). ( -40.60) >DroEre_CAF1 107020 120 - 1 GGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCCGGGAAUGGUUUUAUCGCCAAUGUUCAGGUCCUCGGGAAUGUACCUUCCGAGUAUAGGAACUGGUGGAUACAAGCGCAUCG (((((.((((....((((((.....)))))))))))))))..((.((((((((((((((..((((..((.(((((((.......))))))).))..)))))))).)))).)))).)))). ( -36.70) >DroYak_CAF1 109591 120 - 1 GGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCCGGGAAUGGUCUUAUCGCCGAUGUGCAAGUCCUCGGGAAUGUACCUACCGAGAAUGGGAACUGGAGGAUGCAAACGCAUCG (((((.((((....((((((.....)))))))))))))))......(((......)))((((((...(((((((((......))).((......))......)))))).....)))))). ( -34.10) >consensus GGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCCAGGAAUGGUUUUUUCGCCAAUUUGCAAGUCCUCGGGAAUGUAUCUUCCGAGAAUUGGAACUGGUGGAUGCAAACGCAUCG ....((((((....((((((.....))))))))))))(((((...((((......))))...........(((((((.......)))))))........))))).(((((....))))). (-29.34 = -30.14 + 0.80)

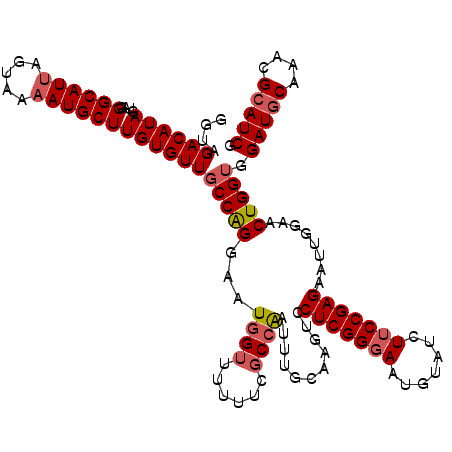
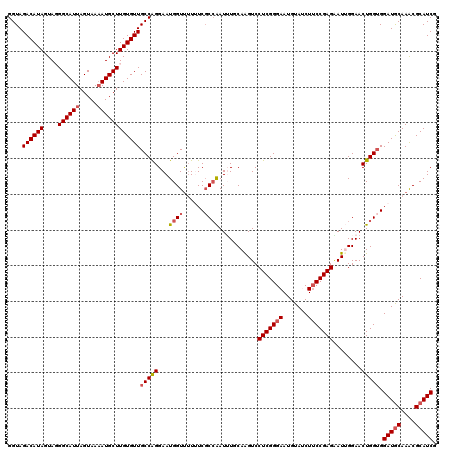
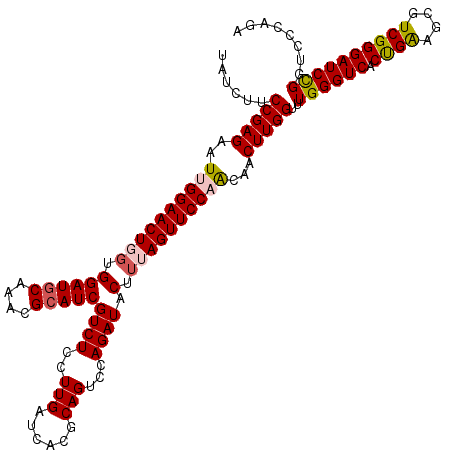
| Location | 7,607,128 – 7,607,248 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607128 120 + 22407834 CAUUCCCGAGGACUUGAAAAUUGGCGAAAUAACCAUUCCUGGCAACACAAGCAUAUUACUAAUGCCCUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCUCUAGUCUUCAAGCC .......(((((((.((...((((.((.(((........((....))...((((.......)))).....))).))..))))(((((((((....))))))))).)).)))))))..... ( -32.30) >DroSec_CAF1 106759 120 + 1 CAUUCCCGAGGACUUGCAAAUUUGCGAAAAAACCAUUCCUGGCAACACAAGCAUUUUACUAAUGCCGUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCCCUCGUUUUCAAGCC ......(((((..(((((....))))).............((((......(((((.....))))).......))))......(((((((((....))))))))).))))).......... ( -31.52) >DroSim_CAF1 112932 120 + 1 CAUUCCCGAGGACCUGCAAAUUGGGGAAAAAACCAUUCCUGGCAACACAAGCAUUUUACUAAUGCCGUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCCCUCGCUUUCAAGCC .......((((.........((((((((.......)))).((((......(((((.....))))).......))))..))))(((((((((....))))))))).))))(((....))). ( -34.62) >DroEre_CAF1 107060 120 + 1 CAUUCCCGAGGACCUGAACAUUGGCGAUAAAACCAUUCCCGGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACCGAGAUCCGGAGUACUUUCCGGAUCCCCUGGUCUUCAAGCC .......(((((((......((((.((((...........(....)....(((((.....))))).......))))..))))(((((((((....)))))))))....)))))))..... ( -35.90) >DroYak_CAF1 109631 120 + 1 CAUUCCCGAGGACUUGCACAUCGGCGAUAAGACCAUUCCCGGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACCGGGAUGCGGAGUACUUUCCGGAUCCCCUGGUCUUCAAGCC ...(((...)))..........(((...(((((((.....((((......(((((.....))))).......))))....(((((.(((((....))))).))))).)))))))...))) ( -36.92) >consensus CAUUCCCGAGGACUUGCAAAUUGGCGAAAAAACCAUUCCUGGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCCCUCGUCUUCAAGCC .......(((((((.(....((((................(....)....(((((.....))))).............))))(((((((((....)))))))))..).)))))))..... (-26.82 = -27.06 + 0.24)

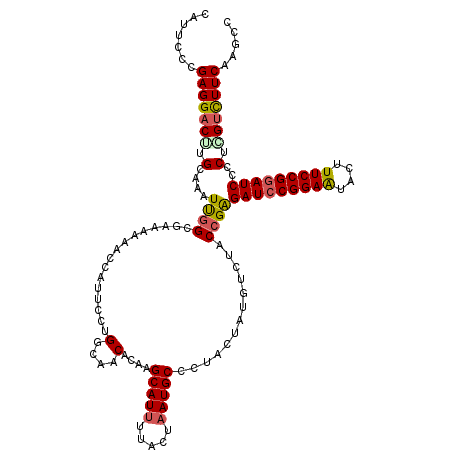
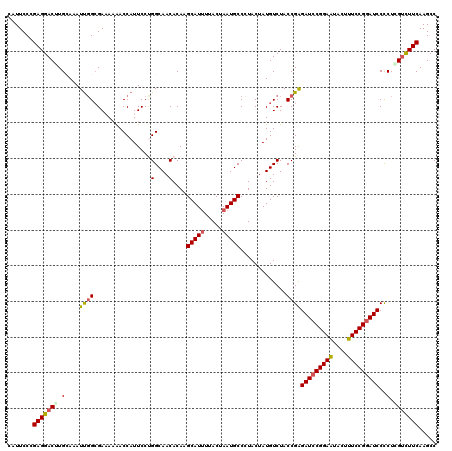
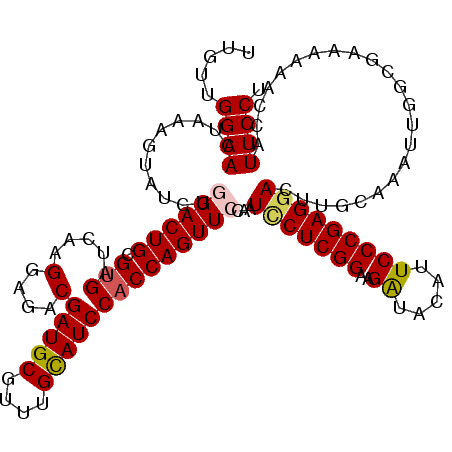
| Location | 7,607,128 – 7,607,248 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -41.76 |
| Consensus MFE | -36.66 |
| Energy contribution | -37.86 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607128 120 - 22407834 GGCUUGAAGACUAGAGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUAGGGCAUUAGUAAUAUGCUUGUGUUGCCAGGAAUGGUUAUUUCGCCAAUUUUCAAGUCCUCGGGAAUG ((((((((((((.(((.((((((((....)))))))))))(((((.((((....(((((.......))))))))))))))))...((((......)))).)))))))))).......... ( -40.90) >DroSec_CAF1 106759 120 - 1 GGCUUGAAAACGAGGGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUACGGCAUUAGUAAAAUGCUUGUGUUGCCAGGAAUGGUUUUUUCGCAAAUUUGCAAGUCCUCGGGAAUG (((((((((((...(((((((((((....)))))))))))(((((.((((....((((((.....))))))))))))))).......))))))..(((....)))))))).......... ( -39.70) >DroSim_CAF1 112932 120 - 1 GGCUUGAAAGCGAGGGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUACGGCAUUAGUAAAAUGCUUGUGUUGCCAGGAAUGGUUUUUUCCCCAAUUUGCAGGUCCUCGGGAAUG ((((((.......((((((((((((....)))))))((..(((((.((((....((((((.....)))))))))))))))..)).........)))))......)))))).......... ( -39.82) >DroEre_CAF1 107060 120 - 1 GGCUUGAAGACCAGGGGAUCCGGAAAGUACUCCGGAUCUCGGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCCGGGAAUGGUUUUAUCGCCAAUGUUCAGGUCCUCGGGAAUG ..(((((.((((..((((((((((......))))))))))(((((.((((....((((((.....))))))))))))))).(((.((((......))))...))).)))).))))).... ( -45.70) >DroYak_CAF1 109631 120 - 1 GGCUUGAAGACCAGGGGAUCCGGAAAGUACUCCGCAUCCCGGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCCGGGAAUGGUCUUAUCGCCGAUGUGCAAGUCCUCGGGAAUG (((...(((((((.......((((......))))..(((((((((.((((....((((((.....))))))))))))))))))).)))))))...)))..........(((...)))... ( -42.70) >consensus GGCUUGAAGACCAGGGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCCAGGAAUGGUUUUUUCGCCAAUUUGCAAGUCCUCGGGAAUG (((..((((((((.((((((((((......))))))))))(((((.((((....((((((.....))))))))))))))).....))))))))..)))..........(((...)))... (-36.66 = -37.86 + 1.20)

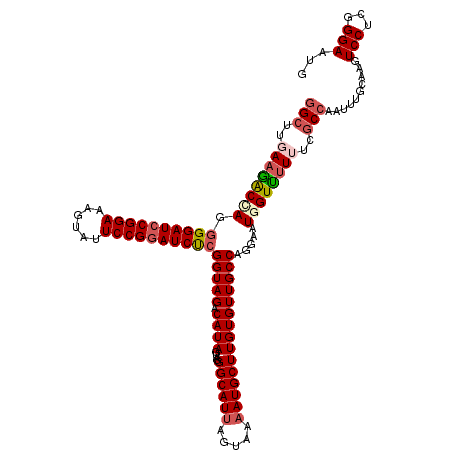
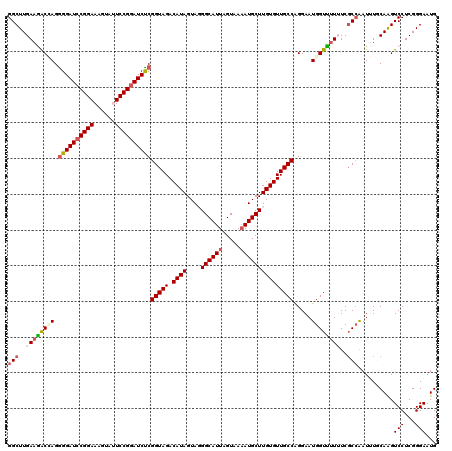
| Location | 7,607,168 – 7,607,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607168 120 + 22407834 GGCAACACAAGCAUAUUACUAAUGCCCUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCUCUAGUCUUCAAGCCGGAACGAUGGAUGGAUAUGAAAACCACCUCGAAUACCCCA (((.......((((.......))))........(.(((..(.(((((((((....))))))))))..))).).....)))....(((.((.(((.........))))))))......... ( -31.10) >DroSec_CAF1 106799 117 + 1 GGCAACACAAGCAUUUUACUAAUGCCGUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCCCUCGUUUUCAAGCCGGAACGAUGGAUGGAUAUGAAAACCACCACGCAUAC---G .((.......(((((.....))))).....(((((((((((.(((((((((....)))))))))...(((((((......)))))))))).))))))))...........))....---. ( -35.00) >DroSim_CAF1 112972 117 + 1 GGCAACACAAGCAUUUUACUAAUGCCGUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCCCUCGCUUUCAAGCCGGAACGAUGGAUGGAUAUGAAAACCACCACGCAUAC---G .((.......(((((.....))))).....(((((((((((.(((((((((....))))))))).((..(((....))).)).....))).))))))))...........))....---. ( -33.40) >DroEre_CAF1 107100 117 + 1 GGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACCGAGAUCCGGAGUACUUUCCGGAUCCCCUGGUCUUCAAGCCGGAGCGAUGGAUGGAUAUGAAAAUGACCUCGCAAAC---A .((.......(((((.....))))).....(((((((((((.(((((((((....)))))))))..(((((......))))).....))).))))))))...........))....---. ( -35.60) >DroYak_CAF1 109671 117 + 1 GGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACCGGGAUGCGGAGUACUUUCCGGAUCCCCUGGUCUUCAAGCCGGAAAGAUGGUUGGAUAUGAAAGCGAGCUCGUAUGC---G .(((......(((((.....))))).....(((((((((((((((.(((((....))))).)))))(((((......)))))......)).))))))))...(((....))).)))---. ( -39.50) >consensus GGCAACACAAGCAUUUUACUAAUGCCCUACUAUGUCUACCGAGAUCCGGAAUACUUUCCGGAUCCCCUCGUCUUCAAGCCGGAACGAUGGAUGGAUAUGAAAACCACCUCGCAUAC___G (....)....(((((.....))))).....(((((((((((.(((((((((....)))))))))..((.((......)).)).....))).))))))))..................... (-27.48 = -27.48 + 0.00)

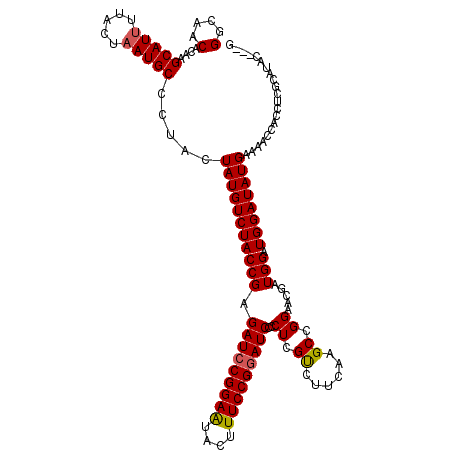
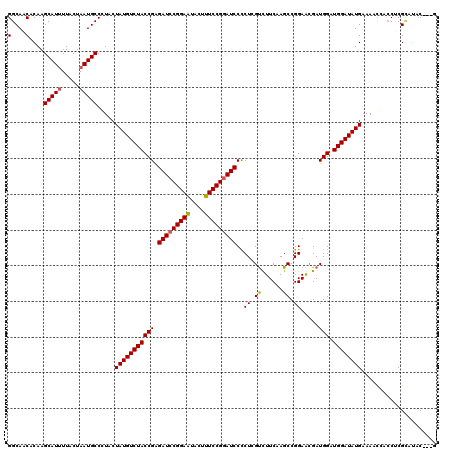
| Location | 7,607,168 – 7,607,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607168 120 - 22407834 UGGGGUAUUCGAGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAGACUAGAGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUAGGGCAUUAGUAAUAUGCUUGUGUUGCC ...(((.((((((.(((.....................))).)))))).))).(((.((((((((....)))))))))))(((((.((((....(((((.......)))))))))))))) ( -36.60) >DroSec_CAF1 106799 117 - 1 C---GUAUGCGUGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAAACGAGGGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUACGGCAUUAGUAAAAUGCUUGUGUUGCC (---(...(((((((((.........))).))).)))..)).((((....))))(((((((((((....)))))))))))(((((.((((....((((((.....))))))))))))))) ( -39.80) >DroSim_CAF1 112972 117 - 1 C---GUAUGCGUGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAAGCGAGGGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUACGGCAUUAGUAAAAUGCUUGUGUUGCC (---(...(((((((((.........))).))).)))..)).((((....))))(((((((((((....)))))))))))(((((.((((....((((((.....))))))))))))))) ( -40.00) >DroEre_CAF1 107100 117 - 1 U---GUUUGCGAGGUCAUUUUCAUAUCCAUCCAUCGCUCCGGCUUGAAGACCAGGGGAUCCGGAAAGUACUCCGGAUCUCGGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCC (---((((((..((((...((((...((............))..))))))))..((((((((((......)))))))))).)))))))..((((.(((..(((.....)))))).)))). ( -37.70) >DroYak_CAF1 109671 117 - 1 C---GCAUACGAGCUCGCUUUCAUAUCCAACCAUCUUUCCGGCUUGAAGACCAGGGGAUCCGGAAAGUACUCCGCAUCCCGGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCC .---((((((((((..(((...((..((.((.((.((((((((.....).))..(((((.((((......)))).))))))).))).)).)).))..)).))).....))))))).))). ( -33.60) >consensus C___GUAUGCGAGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAGACCAGGGGAUCCGGAAAGUAUUCCGGAUCUCGGUAGACAUAGUAGGGCAUUAGUAAAAUGCUUGUGUUGCC ................(((((((...((............))..)))))))...((((((((((......))))))))))(((((.((((....((((((.....))))))))))))))) (-31.50 = -31.98 + 0.48)

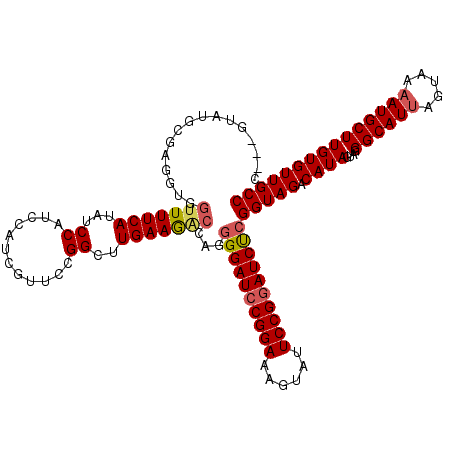
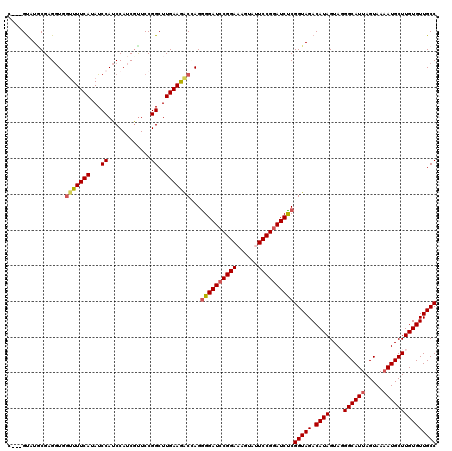
| Location | 7,607,208 – 7,607,328 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -28.68 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7607208 120 - 22407834 AGUUCUUGGGCCCUGAACUAAACGGAAUAUAUGCUAAAGGUGGGGUAUUCGAGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAGACUAGAGGAUCCGGAAAGUAUUCCGGAUCUC (((..(..((((..((((......((((((.(((.....)))..))))))..(((((.............))))))))).))))..)..)))...((((((((((....)))))))))). ( -41.72) >DroSec_CAF1 106839 117 - 1 AGUUCUUGGGCCCUGAACUAAACGGAAUAUAUGCCAAAGGC---GUAUGCGUGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAAACGAGGGGAUCCGGAAAGUAUUCCGGAUCUC .(((.(..((((..((((...(((...((((((((...)))---))))))))(((((.............))))))))).))))..).)))...(((((((((((....))))))))))) ( -45.02) >DroSim_CAF1 113012 117 - 1 AGUUCUUGGGCCCUGAACUGAACGGAAUAUAUGCCAAAGGC---GUAUGCGUGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAAGCGAGGGGAUCCGGAAAGUAUUCCGGAUCUC .(((.(..((((..((((...(((...((((((((...)))---))))))))(((((.............))))))))).))))..).)))...(((((((((((....))))))))))) ( -45.02) >DroEre_CAF1 107140 117 - 1 AGUUCUUGGGUCCAGCACUAAAGGGAAUAUAUGCUAGAGGU---GUUUGCGAGGUCAUUUUCAUAUCCAUCCAUCGCUCCGGCUUGAAGACCAGGGGAUCCGGAAAGUACUCCGGAUCUC .((..(..((((.((((((....((........))...)))---))).((((((................)).))))...))))..)..))...((((((((((......)))))))))) ( -34.59) >DroYak_CAF1 109711 117 - 1 AGUUCUUGGGCCCCGAACUAAAAGGAAUAUAUGCUAGAGGC---GCAUACGAGCUCGCUUUCAUAUCCAACCAUCUUUCCGGCUUGAAGACCAGGGGAUCCGGAAAGUACUCCGCAUCCC .((..(..((((((.........))...(((((..((.(((---........)))..))..)))))..............))))..)..))...(((((.((((......)))).))))) ( -30.40) >consensus AGUUCUUGGGCCCUGAACUAAACGGAAUAUAUGCUAAAGGC___GUAUGCGAGGUGGUUUUCAUAUCCAUCCAUCGUUCCGGCUUGAAGACCAGGGGAUCCGGAAAGUAUUCCGGAUCUC .(((.(..((((..((((......(.((((..(((...)))...)))).)(.(((((.........))))))...)))).))))..).)))...((((((((((......)))))))))) (-28.68 = -29.68 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:21 2006