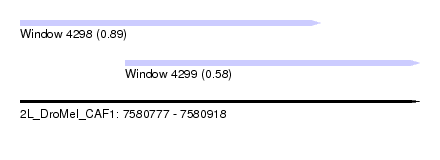
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,580,777 – 7,580,918 |
| Length | 141 |
| Max. P | 0.886990 |

| Location | 7,580,777 – 7,580,883 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -18.91 |
| Energy contribution | -18.94 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7580777 106 + 22407834 CAGUGCCAAUCGAAAGAAGACAGCAAACUGACCGCAG----AGUAGAA----AAAA---AAACUAAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAA-AAACUCUUG-GCU ....(((((((....))...(((....))).((((((----.((((..----....---.........)))).))))..(((((........)))))...))...-......)))-)). ( -26.46) >DroSec_CAF1 80183 106 + 1 CAGUGCCAAUCGAAAGAAGACAGCAAACUGACCGCAG----AGUAGAA----A--A---AAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAAAAAACUCUUGGGUU ((((((...((....))...(((....))).(((..(----(((....----.--.---..))))..(((((.......)))))..)))......))))))......((((....)))) ( -26.00) >DroSim_CAF1 85217 104 + 1 CAGUGCCAAUCGAAAGAAGACAGCAAACUGACCGCAG----AGUAGAA----A--A---AAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAA-AAACUCUUG-GUU ((((((...((....))...(((....))).(((..(----(((....----.--.---..))))..(((((.......)))))..)))......))))))....-.........-... ( -24.90) >DroEre_CAF1 79894 103 + 1 CAGUGCCAAUCGAAAGAAGACAGCAAACUGACCGCGG----AGUAGAA----AAAACU------CGAGCUGCACUGCCCGCAGUCUCGGAUAACUGCACUGAGCA-AAACUCCAU-GCU ((((((...((....))...(((....))).((...(----(((....----...)))------)(((((((.......)))).)))))......))))))((((-........)-))) ( -29.50) >DroYak_CAF1 82098 109 + 1 CAGUGCCAAUCGAAAGAAGACAGCAAACUGACCGCAG----AGCAGAA----AAAACUCAAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGAACG-AAACUCUUU-GCU ((((((...((....)).........((((...((((----.((((..----................)))).))))...))))...........))))))....-.........-... ( -25.97) >DroAna_CAF1 76482 101 + 1 CAGUGCCAAUCGACAGAAGACAGCAGACUGACCGCAGUUGUAGUGAAAAAUAAAAACAAAAGUUAAAACUGCAC--------GUCUCAGAUGACUGCACUGAGGA-AAAU--------- ((((((..(((....((.(((....(((((....)))))(((((.......................)))))..--------))))).)))....))))))....-....--------- ( -21.80) >consensus CAGUGCCAAUCGAAAGAAGACAGCAAACUGACCGCAG____AGUAGAA____AAAA___AAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGAAAA_AAACUCUUG_GCU ((((((...((....))..........((((..((((....((...................))....))))((((....)))).))))......)))))).................. (-18.91 = -18.94 + 0.03)

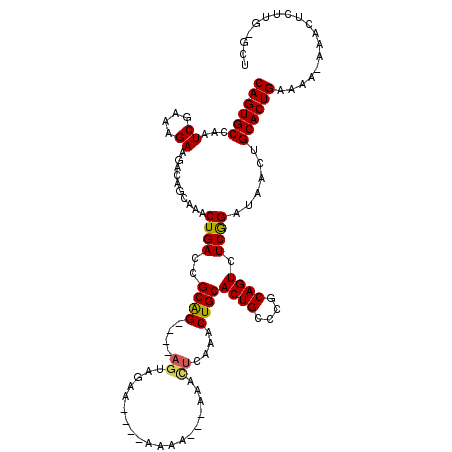
| Location | 7,580,814 – 7,580,918 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -17.14 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7580814 104 + 22407834 AGUAGAAAAAA---AAACUAAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAA-AAACUCUUG-GCUUUUCUAUGUGUUCUUAAAAAUCUGCUCAAG----UAACA ((((((.....---...(..(.(((((.......))))).)..).(((..((((((((((-(..(....)-..))))))).))))..)))....))))))....----..... ( -23.00) >DroSec_CAF1 80220 104 + 1 AGUAGAAA--A---AAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAAAAAACUCUUGGGUUUUUCUAUGUGUUUUUAAGAUUCUGCUCAAG----UUAAA (((((((.--.---.........((((.......))))(((..((.....((((((((((((..(....)..)))))))).)))).))..))))))))))....----..... ( -25.20) >DroSim_CAF1 85254 102 + 1 AGUAGAAA--A---AAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAA-AAACUCUUG-GUUUUUCUGUGUGUUCUUAAAAUUCUGCUCAAG----UAAAA (((((((.--.---...((.(.(((((.......))))).).)).(((..((((((((((-((.(....)-.)))))))).))))..)))...)))))))....----..... ( -21.70) >DroEre_CAF1 79931 104 + 1 AGUAGAAAAAACU------CGAGCUGCACUGCCCGCAGUCUCGGAUAACUGCACUGAGCA-AAACUCCAU-GCUUGUCUAGGUGUUCUUAAAAUUCUGCUCA-GUUUUUUAUU (((((((.....(------((((((((.......)))).))))).(((..((((((((((-........)-)))).....)))))..)))...)))))))..-.......... ( -26.90) >DroYak_CAF1 82135 111 + 1 AGCAGAAAAAACUCAAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGAACG-AAACUCUUU-GCUUUUCUAUGUGUUCUUAAAAUUCUGCUCAAGAUGUUUAUG (((((((..........((.(.(((((.......))))).).)).(((..((((.(((((-((....)))-)...)))...))))..)))...)))))))............. ( -19.10) >consensus AGUAGAAAAAA___AAACUCAAACUGCACUGCCCGCAGUCUCGGAUAACUGCACUGGAAA_AAACUCUUG_GCUUUUCUAUGUGUUCUUAAAAUUCUGCUCAAG____UUAAA (((((((...............(((((.......)))))......(((..((((((((((..............)))))).))))..)))...)))))))............. (-17.14 = -16.90 + -0.24)

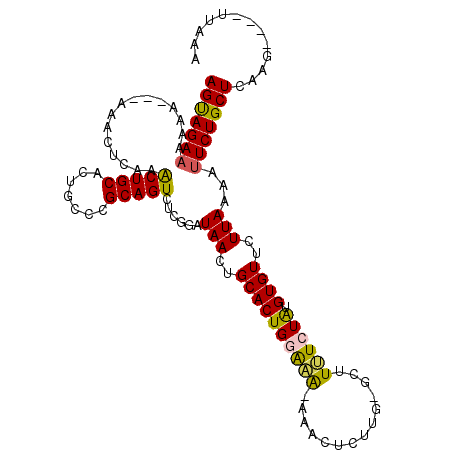
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:06 2006