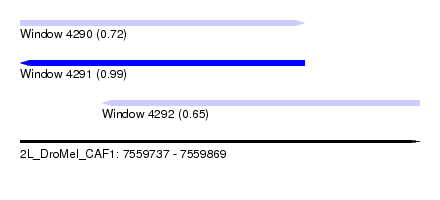
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,559,737 – 7,559,869 |
| Length | 132 |
| Max. P | 0.986665 |

| Location | 7,559,737 – 7,559,831 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -15.14 |
| Consensus MFE | -9.93 |
| Energy contribution | -10.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
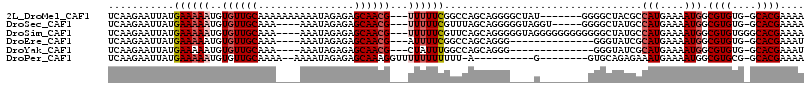
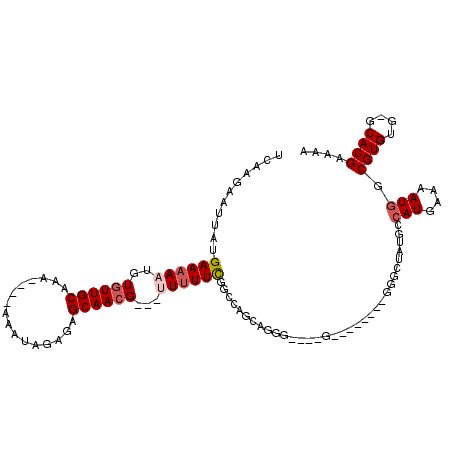
>2L_DroMel_CAF1 7559737 94 + 22407834 UUAAGGUUUUUUUUUUUGUUUUUUACAUUUUCGUGC-CACACGCCAUUUUCAUGGCGUAGCCCC-------AUAGCCCCUGCUGGCCGAAAAACGUUGCUCU .(((.((.........(((.....)))((((((.((-(..(((((((....)))))))......-------..(((....)))))))))))))).))).... ( -22.40) >DroSec_CAF1 59450 83 + 1 -------------UUUUGUAUUUCACAUUUUCGUGC-CACACGCCAUUUUCAUGGCAUAGCCCC-----ACCUACCCCCUGCUAAACGAAAAACGUUGCUCU -------------..............(((((((..-.....(((((....))))).((((...-----...........)))).))))))).......... ( -13.94) >DroSim_CAF1 64160 89 + 1 -------------UUUUGUUUUUUACAUUUUCGUGCCCACACGCCAUUUUCAUGGCAUAGCCCCCCCCCCCCUACCCCCUGCUGAACGAAAAACGUUGCUCU -------------..............(((((((........(((((....))))).((((...................)))).))))))).......... ( -13.31) >DroEre_CAF1 58626 74 + 1 -------------UUCUGUUUUUCACAAUUUCGUGC-CACACGCCAUUUUCAUGCGAUACCC--------------CCCUGCUGGCCGAAAAUCGUUGCUCU -------------...............(((((.((-(((((((.........)))......--------------...)).)))))))))........... ( -10.90) >consensus _____________UUUUGUUUUUCACAUUUUCGUGC_CACACGCCAUUUUCAUGGCAUAGCCCC_______CUACCCCCUGCUGAACGAAAAACGUUGCUCU .................(((((((........(((....)))(((((....)))))...............................)))))))........ ( -9.93 = -10.68 + 0.75)


| Location | 7,559,737 – 7,559,831 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -15.02 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7559737 94 - 22407834 AGAGCAACGUUUUUCGGCCAGCAGGGGCUAU-------GGGGCUACGCCAUGAAAAUGGCGUGUG-GCACGAAAAUGUAAAAAACAAAAAAAAAAACCUUAA ......(((((((.((((((.....((((..-------..))))(((((((....))))))).))-)).)))))))))........................ ( -28.80) >DroSec_CAF1 59450 83 - 1 AGAGCAACGUUUUUCGUUUAGCAGGGGGUAGGU-----GGGGCUAUGCCAUGAAAAUGGCGUGUG-GCACGAAAAUGUGAAAUACAAAA------------- ..(((..((..(.((.((.....)).)).)..)-----)..)))..(((((....)))))((((.-.(((......)))..))))....------------- ( -22.30) >DroSim_CAF1 64160 89 - 1 AGAGCAACGUUUUUCGUUCAGCAGGGGGUAGGGGGGGGGGGGCUAUGCCAUGAAAAUGGCGUGUGGGCACGAAAAUGUAAAAAACAAAA------------- ...((.((((((..(.(((.((.....))...))).)..)))).(((((((....)))))))))..)).....................------------- ( -21.30) >DroEre_CAF1 58626 74 - 1 AGAGCAACGAUUUUCGGCCAGCAGGG--------------GGGUAUCGCAUGAAAAUGGCGUGUG-GCACGAAAUUGUGAAAAACAGAA------------- ......(((((((.((((((.((.((--------------(....)).(((....))).).))))-)).)))))))))...........------------- ( -16.50) >consensus AGAGCAACGUUUUUCGGCCAGCAGGGGGUAG_______GGGGCUAUGCCAUGAAAAUGGCGUGUG_GCACGAAAAUGUAAAAAACAAAA_____________ ......(((((((.((.((....))..................((((((((....))))))))......)))))))))........................ (-15.02 = -14.52 + -0.50)



| Location | 7,559,764 – 7,559,869 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -12.76 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7559764 105 - 22407834 UCAAGAAUUAUGAAAAAUGUGUUGCAAAAAAAAAAUAGAGAGCAACG---UUUUUCGGCCAGCAGGGGCUAU-------GGGGCUACGCCAUGAAAAUGGCGUGUG-GCACGAAAA .................(((((..(...............(((..((---(.....((((......))))))-------)..)))(((((((....))))))))..-))))).... ( -32.30) >DroSec_CAF1 59464 103 - 1 UCAAGAAUUAUGAAAAAUGUGUUGCAAA----AAAUAGAGAGCAACG---UUUUUCGUUUAGCAGGGGGUAGGU-----GGGGCUAUGCCAUGAAAAUGGCGUGUG-GCACGAAAA ((..(....(((((((((((..(((...----.........))))))---))))))))....).........((-----(..((.(((((((....))))))))).-.)))))... ( -26.80) >DroSim_CAF1 64174 109 - 1 UCAAGAAUUAUGAAAAAUGUGUUGCAAA----AAAUAGAGAGCAACG---UUUUUCGUUCAGCAGGGGGUAGGGGGGGGGGGGCUAUGCCAUGAAAAUGGCGUGUGGGCACGAAAA .................(((((..(...----........(((..(.---(((..((..(......)..)..)..))).)..)))(((((((....))))))))..).)))).... ( -25.40) >DroEre_CAF1 58640 94 - 1 UCAAGAAUUAUGAAAAAUGUGUUGCAAA----AAAUAGAGAGCAACG---AUUUUCGGCCAGCAGGG--------------GGGUAUCGCAUGAAAAUGGCGUGUG-GCACGAAAU .................(((((..((..----.....(((((.....---.))))).((((.(((.(--------------(....)).).))....)))).))..-))))).... ( -19.80) >DroYak_CAF1 60723 94 - 1 UCAAGAAUUAUGAAAAAUGUGUUGCAAA----AAAUAGAGAGCAACG---CUAUUUGGCCAGCAGGG--------------GGGUAUCGCAUGAAAAUGGCGUGUG-GCACGAAAU .................(((((..((..----((((((.(.....).---)))))).((((.(((.(--------------(....)).).))....)))).))..-))))).... ( -20.70) >DroPer_CAF1 80644 94 - 1 UCAAGAAUUAUGAAAAAUGUGUUGCAAAA--AAAAUAGAGAGCAAAGGUUUUUUUUUUU-A----------G--------GUGCAGAGAAAUGAAAAUGGCGUGCG-GCACGAAAA .................(((((((((...--((((.(((((((....))))))).))))-.----------.--------.......(..((....))..).))))-))))).... ( -17.10) >consensus UCAAGAAUUAUGAAAAAUGUGUUGCAAA____AAAUAGAGAGCAACG___UUUUUCGGCCAGCAGGG____G________GGGCUAUGCCAUGAAAAUGGCGUGUG_GCACGAAAA ...........((((((..((((((................))))))...)))))).................................(((....))).((((....)))).... (-12.76 = -13.37 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:00 2006