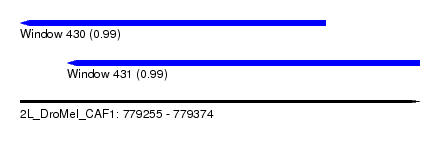
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 779,255 – 779,374 |
| Length | 119 |
| Max. P | 0.992199 |

| Location | 779,255 – 779,346 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 60.62 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -18.17 |
| Energy contribution | -25.55 |
| Covariance contribution | 7.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
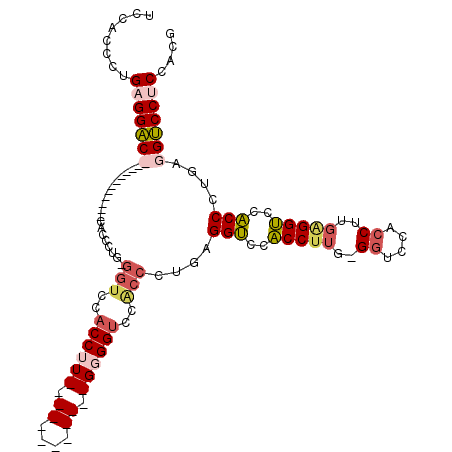
>2L_DroMel_CAF1 779255 91 - 22407834 CCUUGAGGUCCACCCUGGGGUCCGCCCUGUGGUCCACCUUGAGGUCCACCCUGAGGUCCACCCUGAGGUCCGCCCUGGGGUCCACCUUGAG-------------------------- .((..((((..((((..((((..((((.((((...(((((..((....))..)))))))))...).)))..))))..))))..))))..))-------------------------- ( -44.70) >DroSec_CAF1 39666 117 - 1 CCUUGAGGUCCACCCUGGGGUCCACCCUGAGGUCCACCUUGGGGUCCACCUUGGGGUCCACCUUGGGGUCCACCUUGGGGUCCACCUUGGGGUCCACCUUGGGGUCCACCAUGGGGU (((((.(((..((((..((((..((((..((((..((((..((((..((((..(((....)))..))))..))))..))))..))))..))))..))))..))))..))).))))). ( -69.50) >DroEre_CAF1 39309 79 - 1 CCAU------------GAGGUCCACCAUGAGGUCCACCCUGCGCUCCACCAUGAGGUCCACCCUGAGGACCUCCACGUCGGCCACCUUGAG-------------------------- ....------------..((((.((...(((((((.(..((.((..(.....)..)).))....).)))))))...)).))))........-------------------------- ( -21.20) >DroAna_CAF1 40313 70 - 1 CCU---------------GGUCCACCCUGAGGACCUCCU---GGUCCACCUCGAGGGCCUCCU---GGUCCUCCACGACGGCCACCCUGAG-------------------------- ..(---------------((((........(((((....---))))).....(((((((....---)))))))......))))).......-------------------------- ( -28.10) >consensus CCUU____________GGGGUCCACCCUGAGGUCCACCUUG_GGUCCACCUUGAGGUCCACCCUGAGGUCCUCCACGGCGGCCACCUUGAG__________________________ .((((((((..((((((((((..(((.((((((..(((((..((....))..)))))..)))))).)))..))))))))))..)))))))).......................... (-18.17 = -25.55 + 7.38)


| Location | 779,269 – 779,374 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.31 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -19.14 |
| Energy contribution | -21.45 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 779269 105 - 22407834 UCCUCCCUGAGGAC------------CACCCUGGGGUCCACCUUGAGGUCCACCCUGGGGUCCGCCCUGUGGUCCACCUUGAGGUCCACCCUGAGGUCCACCCUGAGGUCCGCCCUG .....(((.(((..------------.(((..(((((..((((..((((..(((.(((((....))))).)))..))))..))))..)))))..)))....))).)))......... ( -46.60) >DroSec_CAF1 39706 117 - 1 UCCGCCCUGAGGACCGCCCUGAGGACCGCCCUGUGGUCCACCUUGAGGUCCACCCUGGGGUCCACCCUGAGGUCCACCUUGGGGUCCACCUUGGGGUCCACCUUGGGGUCCACCUUG ........((((..........(((((((...)))))))((((..((((..((((..((((..((((..(((....)))..))))..))))..))))..))))..))))...)))). ( -61.70) >DroEre_CAF1 39323 105 - 1 UCCACCAUGAGGUCCAUCGUGCGCUCCACCAUGAGGUCCACCAU------------GAGGUCCACCAUGAGGUCCACCCUGCGCUCCACCAUGAGGUCCACCCUGAGGACCUCCACG ........(((((((.(((.((((.....((((.((.((.....------------..)).))..))))(((.....))))))).).......(((.....)))))))))))).... ( -31.70) >DroAna_CAF1 40327 81 - 1 ACCACCCUGAGGAC------------CGCCUGG---GCCUCCU---------------GGUCCACCCUGAGGACCUCCU---GGUCCACCUCGAGGGCCUCCU---GGUCCUCCACG ........((((..------------....(((---(((....---------------))))))......(((((....---))))).))))(((((((....---))))))).... ( -37.80) >consensus UCCACCCUGAGGAC____________CACCCUG_GGUCCACCUU____________GGGGUCCACCCUGAGGUCCACCUUG_GGUCCACCUUGAGGUCCACCCUGAGGUCCUCCACG ........(((((((...................(((..((((((.((....)).))))))..)))....(((..(((((..((....))..)))))..)))....))))))).... (-19.14 = -21.45 + 2.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:40 2006