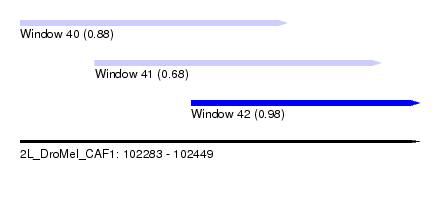
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 102,283 – 102,449 |
| Length | 166 |
| Max. P | 0.983180 |

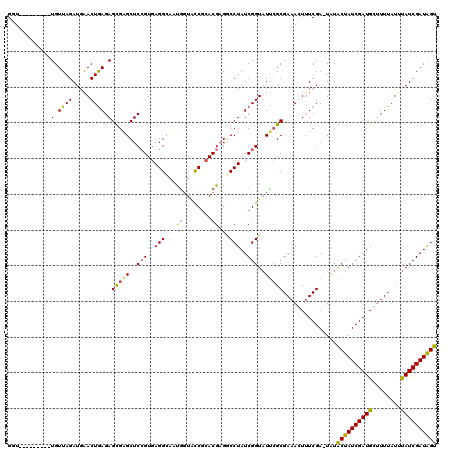
| Location | 102,283 – 102,394 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 102283 111 + 22407834 GGU---------UGUUAGAUUGACUAAGAGUGAGCUCCGUGAGGCAAUGGUAUCGAACGCGACCUAUCGGUAUUCGCGAAACUUUCGAGUAUACUAUCGAUGUUUUUAUUUAUCGAUAGU (((---------(((..(((..((((.(((....)))(....)....)))))))....)))))).....(((((((.........)))))))((((((((((........)))))))))) ( -29.00) >DroSec_CAF1 2070 110 + 1 GUU---------UGUUAGAUGAACUGAGAGCGAGCUCAGUGAGGCAAUGGUACCGCACGAGCCCUAUCGGUAUCCGCGAAAGUUUCGA-UAUGCUAUCGAUGCUUUUAUUUAUCGAUAGU (((---------(.((((.....))))))))..((((.(((.((........)).))))))).(((((((((.....((((((.((((-((...)))))).))))))...))))))))). ( -33.30) >DroSim_CAF1 2070 110 + 1 GUU---------UGUUAGAUGAACUGAGAGCGAGCUCCGUGAGGCAAUGGUACCGCACGAGGCCUAUCGGUAUUCGCGAAAGUUUCGA-UAUGCUAUCGAUGCUUUUAUUUAUCGAUAGU (((---------(.((((.....))))))))..((..((((.((........)).))))..))(((((((((.....((((((.((((-((...)))))).))))))...))))))))). ( -34.20) >DroYak_CAF1 7152 120 + 1 GGUCCUCUUUGGUGUUAGAUGAACUGAGAGUGGGCUCCGUGAGGUAUCAUUGCCUCACGAGGCCUAUCGGUUUUAGCGAAACUAUCGAGUGUACUAUCGAUGUCUUUAUUCAUCGAUGGU .(..(((..((((....(..(((((((...(((((..(((((((((....)))))))))..))))))))))))...)...))))..)))..)((((((((((........)))))))))) ( -50.90) >consensus GGU_________UGUUAGAUGAACUGAGAGCGAGCUCCGUGAGGCAAUGGUACCGCACGAGGCCUAUCGGUAUUCGCGAAACUUUCGA_UAUACUAUCGAUGCUUUUAUUUAUCGAUAGU ............(.((((.....)))).)(((((..(((..((((..((........))..))))..)))..)))))...............((((((((((........)))))))))) (-22.96 = -23.03 + 0.06)

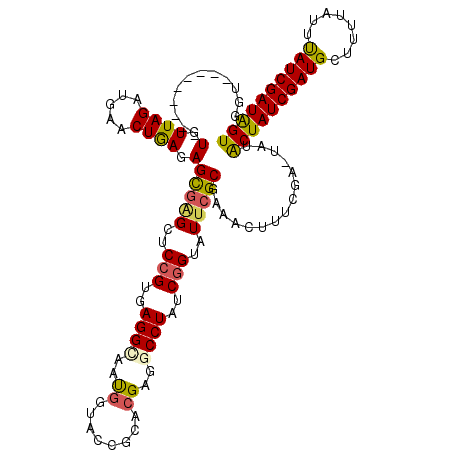
| Location | 102,314 – 102,433 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -24.29 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 102314 119 + 22407834 GAGGCAAUGGUAUCGAACGCGACCUAUCGGUAUUCGCGAAACUUUCGAGUAUACUAUCGAUGUUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAGUUUGCCGG ..(((((.(.((((...(((((((....))...)))))........((((((((((((((((........))))))))))))((............))...)))).)))).)))))).. ( -35.60) >DroSec_CAF1 2101 118 + 1 GAGGCAAUGGUACCGCACGAGCCCUAUCGGUAUCCGCGAAAGUUUCGA-UAUGCUAUCGAUGCUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAAUUUGCCGG ..(((((.(....)((.(((.(.(((((((((.....((((((.((((-((...)))))).))))))...))))))))).))))))..........................))))).. ( -32.30) >DroSim_CAF1 2101 118 + 1 GAGGCAAUGGUACCGCACGAGGCCUAUCGGUAUUCGCGAAAGUUUCGA-UAUGCUAUCGAUGCUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAAUUUGCCGG ..(((((.(....)((.(((.((.((((((((.....((((((.((((-((...)))))).))))))...)))))))))).)))))..........................))))).. ( -33.50) >DroYak_CAF1 7192 119 + 1 GAGGUAUCAUUGCCUCACGAGGCCUAUCGGUUUUAGCGAAACUAUCGAGUGUACUAUCGAUGUCUUUAUUCAUCGAUGGUGUCGGCUUCAAUUACACGUAAACUCAGAUAAUUCGCCGG ((((((....))))))..((((((....)))))).(((((..((((((((.(((((((((((........))))))))((((...........))))))).)))).)))).)))))... ( -38.60) >consensus GAGGCAAUGGUACCGCACGAGGCCUAUCGGUAUUCGCGAAACUUUCGA_UAUACUAUCGAUGCUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAAUUUGCCGG ..(((((.(((((((............)))))))..........((((...(((((((((((........)))))))))))))))...........................))))).. (-24.29 = -23.98 + -0.31)

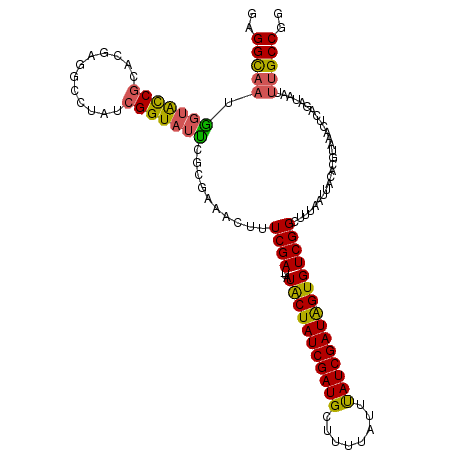
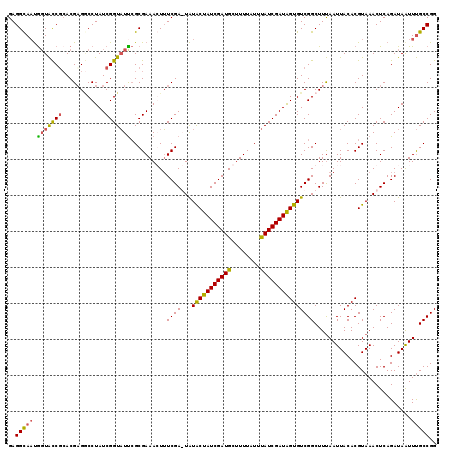
| Location | 102,354 – 102,449 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -21.15 |
| Energy contribution | -20.15 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 102354 95 + 22407834 ACUUUCGAGUAUACUAUCGAUGUUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAGUUUGCCGGUUAAA-AAGACAAUAAA ............((((((((((........))))))))))(((...(((((((....(((((((.....))))))).)))))))-..)))...... ( -22.20) >DroSec_CAF1 2141 94 + 1 AGUUUCGA-UAUGCUAUCGAUGCUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAAUUUGCCGGCUAAA-AAGGCAAUAAA .((((...-...((((((((((........))))))))))((((((...(((((...(......)...))))).))))))....-.))))...... ( -22.00) >DroSim_CAF1 2141 94 + 1 AGUUUCGA-UAUGCUAUCGAUGCUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAAUUUGCCGGCUAAA-AAGGCAAUAAA .((((...-...((((((((((........))))))))))((((((...(((((...(......)...))))).))))))....-.))))...... ( -22.00) >DroYak_CAF1 7232 96 + 1 ACUAUCGAGUGUACUAUCGAUGUCUUUAUUCAUCGAUGGUGUCGGCUUCAAUUACACGUAAACUCAGAUAAUUCGCCGGCUAAAAAAGGCCACAAA ..((((((((.(((((((((((........))))))))((((...........))))))).)))).)))).......((((......))))..... ( -25.50) >consensus ACUUUCGA_UAUACUAUCGAUGCUUUUAUUUAUCGAUAGUGUCGGCUUUAAUUACACGUAAACUCAGAUAAUUUGCCGGCUAAA_AAGGCAAUAAA ............((((((((((........))))))))))((((((...(((((...(......)...))))).))))))................ (-21.15 = -20.15 + -1.00)

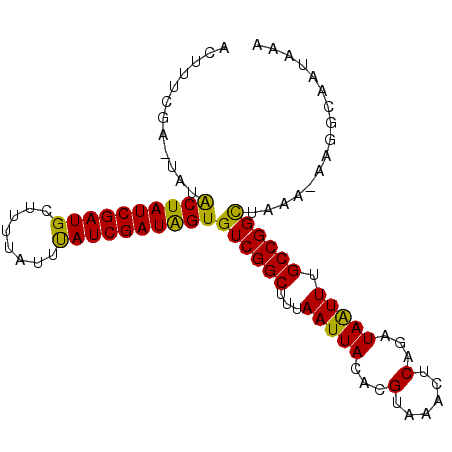
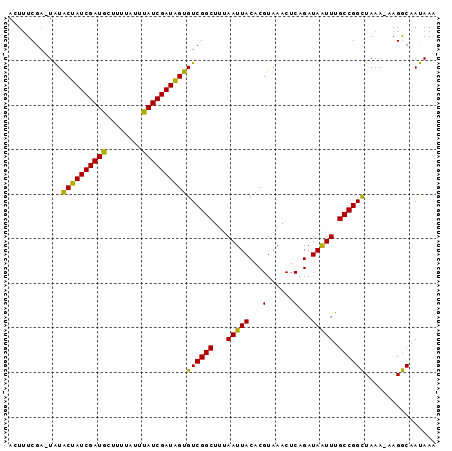
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:38 2006