
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 769,331 – 769,423 |
| Length | 92 |
| Max. P | 0.813354 |

| Location | 769,331 – 769,423 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
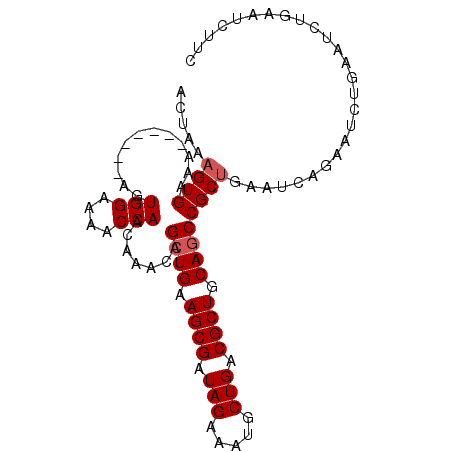
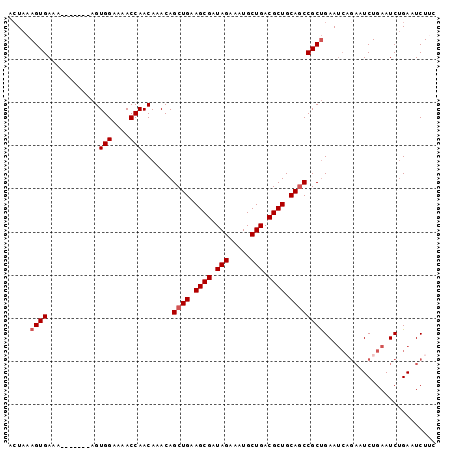
>2L_DroMel_CAF1 769331 92 + 22407834 ACUAAAGUGAAA-------AGUGGAAAACCAACAAACAGCUGAAGCGAUAGAAAUGCUGACGCUGCAGCCGCUGAAUCUGAAUCUGAAUCUGAAUCUUC .....((((...-------..(((....))).......((((.((((.(((.....))).)))).))))))))((.((.((.......)).)).))... ( -18.50) >DroSec_CAF1 30996 86 + 1 ACUAAAGUGAAA-------AGUGGAAAACCAACAAACAGCUGAAGCGAUAGAAAUGCUGACGCUGCACCCGCUGAA------UCUGAAUCUGAAUCUUC ............-------..(((....))).....((((((.((((.(((.....))).)))).))...))))..------................. ( -13.50) >DroSim_CAF1 34420 92 + 1 ACUAAAGUGAAA-------AGUGGAAAACCAACAAACAGCUGAAGCGAUAGAAAUGCUGACGCUGCAGCCGCUGAAUCAGAAUCUGAAUCUGAAUCUUC .....((((...-------..(((....))).......((((.((((.(((.....))).)))).))))))))((.(((((.......))))).))... ( -23.10) >DroEre_CAF1 31216 95 + 1 --UAAAGUGAAAAGUGGAAAGUGGAAAACCAACAAACAGCUGAAGCGAUAGAAAUGCUGACGCUGCAGCCGCCGAAUCAGAAUCUGAAUCUGAAUCU-- --....(((.....((.....(((....))).....))((((.((((.(((.....))).)))).))))))).((.(((((.......))))).)).-- ( -22.90) >DroYak_CAF1 34588 97 + 1 ACUGAAGUGAAAAGUGGAAAGUGGAAAACCAACAAACAGCUGAAGCGAUAGAAAUGCUGACGCUGCAGCCGCUGAAUCAGAAUCUGAAUCAGAAUCU-- .((((((((.....((.....(((....))).....))((((.((((.(((.....))).)))).))))))))...((((...)))).)))).....-- ( -24.60) >consensus ACUAAAGUGAAA_______AGUGGAAAACCAACAAACAGCUGAAGCGAUAGAAAUGCUGACGCUGCAGCCGCUGAAUCAGAAUCUGAAUCUGAAUCUUC .....((((............(((....))).......((((.((((.(((.....))).)))).)))))))).......................... (-15.68 = -16.08 + 0.40)

| Location | 769,331 – 769,423 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
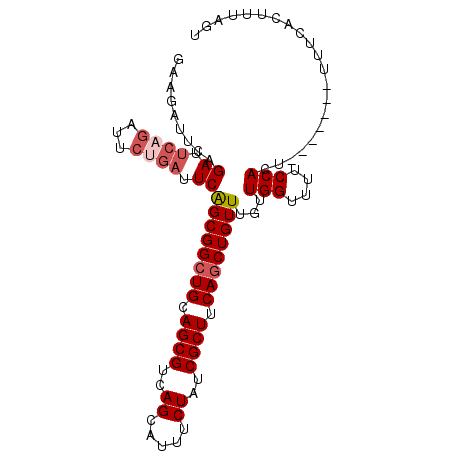
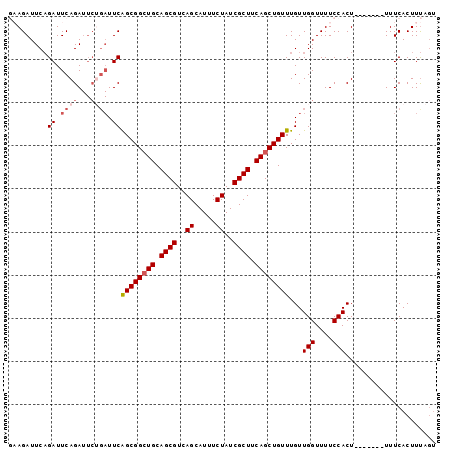
>2L_DroMel_CAF1 769331 92 - 22407834 GAAGAUUCAGAUUCAGAUUCAGAUUCAGCGGCUGCAGCGUCAGCAUUUCUAUCGCUUCAGCUGUUUGUUGGUUUUCCACU-------UUUCACUUUAGU (((((....((.((.......)).))((((((((.((((..((.....))..)))).))))))))...(((....))).)-------))))........ ( -21.50) >DroSec_CAF1 30996 86 - 1 GAAGAUUCAGAUUCAGA------UUCAGCGGGUGCAGCGUCAGCAUUUCUAUCGCUUCAGCUGUUUGUUGGUUUUCCACU-------UUUCACUUUAGU (((((....((.((((.------...(((((.((.((((..((.....))..)))).)).)))))..))))...))...)-------))))........ ( -16.50) >DroSim_CAF1 34420 92 - 1 GAAGAUUCAGAUUCAGAUUCUGAUUCAGCGGCUGCAGCGUCAGCAUUUCUAUCGCUUCAGCUGUUUGUUGGUUUUCCACU-------UUUCACUUUAGU (((((.(((((.......))))).))((((((((.((((..((.....))..)))).))))))))...(((....)))..-------.)))........ ( -26.10) >DroEre_CAF1 31216 95 - 1 --AGAUUCAGAUUCAGAUUCUGAUUCGGCGGCUGCAGCGUCAGCAUUUCUAUCGCUUCAGCUGUUUGUUGGUUUUCCACUUUCCACUUUUCACUUUA-- --.((.(((((.......))))).))((((((((.((((..((.....))..)))).))))))))...(((....)))...................-- ( -25.50) >DroYak_CAF1 34588 97 - 1 --AGAUUCUGAUUCAGAUUCUGAUUCAGCGGCUGCAGCGUCAGCAUUUCUAUCGCUUCAGCUGUUUGUUGGUUUUCCACUUUCCACUUUUCACUUCAGU --.....((((.((((...)))).))))((((((.((((..((.....))..)))).)))))).....(((....)))..................... ( -22.90) >consensus GAAGAUUCAGAUUCAGAUUCUGAUUCAGCGGCUGCAGCGUCAGCAUUUCUAUCGCUUCAGCUGUUUGUUGGUUUUCCACU_______UUUCACUUUAGU .........((.((((...)))).))((((((((.((((..((.....))..)))).))))))))...(((....)))..................... (-17.74 = -18.78 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:38 2006