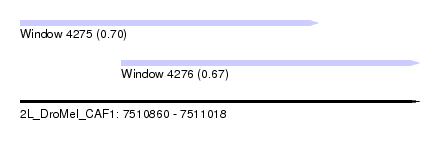
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,510,860 – 7,511,018 |
| Length | 158 |
| Max. P | 0.702642 |

| Location | 7,510,860 – 7,510,978 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.35 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.85 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
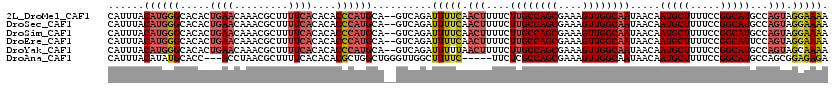
>2L_DroMel_CAF1 7510860 118 + 22407834 CAUUUACAUGGGCACACUGAACAAACGCUUUUCACACACCCAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAA ......((((((.....((((.........))))....))))))..--......(((((.(((....((((((((....)))))))).....(((((.....)))))...))).))))). ( -30.50) >DroSec_CAF1 13073 118 + 1 CAUUUACAUGGGCACACUGAACAAACGCUUUUCACACACCCAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAA ......((((((.....((((.........))))....))))))..--......(((((.(((....((((((((....)))))))).....(((((.....)))))...))).))))). ( -30.50) >DroSim_CAF1 16221 118 + 1 CAUUUACAUGGGCACACUGAACAAACGCUUUUCACACACCCAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAA ......((((((.....((((.........))))....))))))..--......(((((.(((....((((((((....)))))))).....(((((.....)))))...))).))))). ( -30.50) >DroEre_CAF1 11916 118 + 1 CAUUUACAUGGGCACACUGAACAAACGCUUUUCACACACCCAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUUCCAGUAGGAAAA ......((((((.....((((.........))))....))))))..--......(((((.(((....((((((((....))))))))....((((((.....))))))..))).))))). ( -30.80) >DroYak_CAF1 12459 118 + 1 CAUUUACAUGGGCACACUGAACAAACGCUUUUCACACACCCAUGCA--GUCAGAUUUUUAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGCAAAA ......((((((.....((((.........))))....))))))..--...................((((((((....))))))))......((((...(.((....)).).))))... ( -29.00) >DroAna_CAF1 13451 112 + 1 CAUUUACAUAUGCACC---ACCUAACGCUUUUCACACACGCUGGCUGGGUUGGCUUUUC-----UUCUCGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGCGGAGAGA ................---........((((((.....(((((((.(((..((....))-----..))))))))))...(((((((.......((((.....))))))))))))))))). ( -33.91) >consensus CAUUUACAUGGGCACACUGAACAAACGCUUUUCACACACCCAUGCA__GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAA ......((((((.....((((.........))))....))))))..........(((((.(((....((((((((....)))))))).....(((((.....)))))...))).))))). (-24.82 = -25.85 + 1.03)
| Location | 7,510,900 – 7,511,018 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
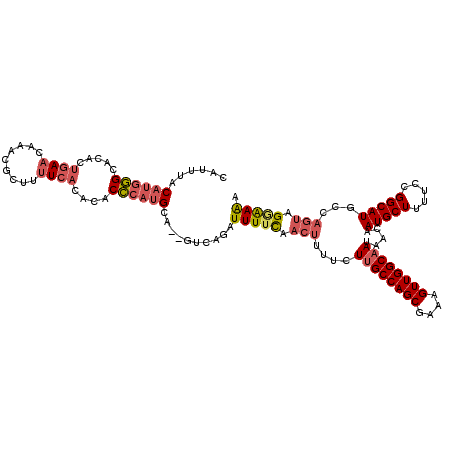
>2L_DroMel_CAF1 7510900 118 + 22407834 CAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAAGAGAUAGCCAUAGAUUGACAGCUGGCAGCAGAGUGAGAUG ..(((.--(((((....((((((....((((((((....))))))))........(((((((.((......)).)))))))..........)).))))...))))).))).......... ( -36.00) >DroSec_CAF1 13113 118 + 1 CAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAAGAGAUAGCCAUAGACUGACAGCUGGCAGCAGAGUGAGAUG ..(((.--(((((....(((.......((((((((....))))))))........(((((((.((......)).)))))))..............)))...))))).))).......... ( -35.20) >DroSim_CAF1 16261 118 + 1 CAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAAGAGAUAGCCAUAGAUUGACAGCUGGCAGCAGAGUGAGAUG ..(((.--(((((....((((((....((((((((....))))))))........(((((((.((......)).)))))))..........)).))))...))))).))).......... ( -36.00) >DroEre_CAF1 11956 118 + 1 CAUGCA--GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUUCCAGUAGGAAAAGAGAUAGCCAUAGAUUGACAGCUGGCAGCAGAGAGAGAUG ..(((.--(((((....((((((....((((((((....))))))))........(((((((.((......)).)))))))..........)).))))...))))).))).......... ( -36.00) >DroYak_CAF1 12499 118 + 1 CAUGCA--GUCAGAUUUUUAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGCAAAAGAGAUAGCCAUAGAUUGACAGCUGGCAGCAGAGAGAGAUG ......--...................((((((((....))))))))........((((..(.((.(((((((.((((................))).).))))))))).)..))))... ( -32.59) >DroAna_CAF1 13488 115 + 1 CUGGCUGGGUUGGCUUUUC-----UUCUCGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGCGGAGAGAGAGACAGCCUCAGAUUGACAGCUGGCAGGCGAUAGAGAUA (((((((((..(((.....-----.....((((((....)))))).........)))..)))))).((((((((..((..(((.....)))...))..).))))))).....)))..... ( -43.11) >consensus CAUGCA__GUCAGAUUUUCAACUUUUCUUGCCAGCGAAAGUUGGCAAUAACAAUGCUUUUCCGGCAUGCCAGUAGGAAAAGAGAUAGCCAUAGAUUGACAGCUGGCAGCAGAGAGAGAUG ..(((...(((................((((((((....))))))))........(((((((.((......)).))))))).))).((((............)))).))).......... (-30.14 = -30.00 + -0.14)

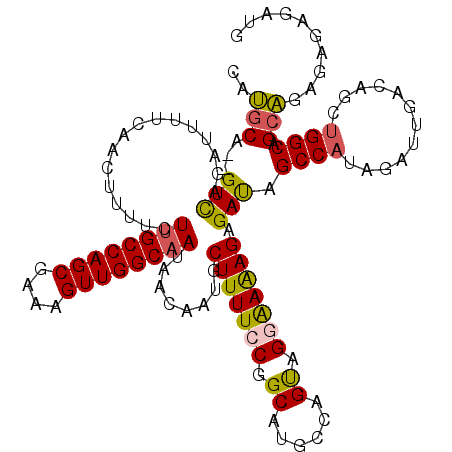

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:45 2006