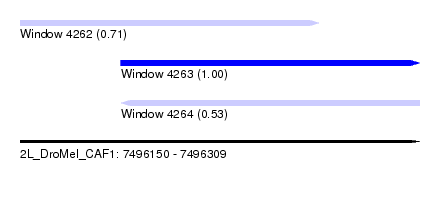
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,496,150 – 7,496,309 |
| Length | 159 |
| Max. P | 0.999823 |

| Location | 7,496,150 – 7,496,269 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -31.60 |
| Energy contribution | -32.08 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7496150 119 + 22407834 AUUAUCUGGUUGAUUACUCGCAAUUUACGUGCGUUUGGCGUUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA-UUGGCGUGGAGUUUGGCUUUUCGAAUUGCCGCUA ...................((((...(((..((..((.(......).))..))..)))))))((((((((((((((((((((((.-...)))....)))))....)))))))))))))). ( -35.80) >DroSec_CAF1 18894 119 + 1 AUUAUCUGGUUGAUUACUCGCAAUUUACGUGCGUUUGGCGUUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA-UUGGCGUGGAGUUUGUCUUUUCGAAUUGCCGCUA ...................((((...(((..((..((.(......).))..))..)))))))((((((((((((((((((((((.-...)))....))))))....))))))))))))). ( -36.60) >DroSim_CAF1 18958 119 + 1 AUUAUCUGGUUGAUUACUCGCAAUUUACGUGCGUUAGGCGUUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA-UUGGCGUGGAGUUUGUCUUUUUGAAUUGCCGCUA ...................((((...(((..((...(.(......).)...))..)))))))((((((((((..((((((((((.-...)))....))))))....)..)))))))))). ( -33.70) >DroEre_CAF1 19222 119 + 1 AUUAUCUAGUUGAUUACUCGUAAUUUACGUGCGUUUGCCGUUUGAGGUGUUUGUGCGUUUGCAGCGGCGACUUGAACGAACCGUA-CUGGCGUGGAGUUUAUGUUUUCGAAUUGCCGCCA ...................((((...(((..((..((((......))))..))..))))))).(((((((.(((((.((((..((-(....)))..)))).....))))).))))))).. ( -32.10) >DroYak_CAF1 19366 120 + 1 AUUAUCUGCUUGAUUACUCGCAAUUUACGUGCGUUUGGCGUUUGAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGUACCGUUUUUGGCGUGGAGUUUAUCUUUUCGAAUUGCCGCUA ...................((((...(((..((..((.(......).))..))..)))))))((((((((((((((((..(((....)))..))(((.....))))))))))))))))). ( -36.60) >consensus AUUAUCUGGUUGAUUACUCGCAAUUUACGUGCGUUUGGCGUUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA_UUGGCGUGGAGUUUGUCUUUUCGAAUUGCCGCUA ...................((((...(((..((..((.(......).))..))..)))))))((((((((((((((((..(((....)))..))(((.....))))))))))))))))). (-31.60 = -32.08 + 0.48)

| Location | 7,496,190 – 7,496,309 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -47.02 |
| Consensus MFE | -42.78 |
| Energy contribution | -43.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7496190 119 + 22407834 UUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA-UUGGCGUGGAGUUUGGCUUUUCGAAUUGCCGCUAACAGAGCGCUGAGCUCCUCACGCGAACGAGAUCCACCGAC .....((((..(.(.(((((((((((((((((((((((((((((.-...)))....)))))....))))))))))))))....((((.....)))).....)))))))).)..))))... ( -47.00) >DroSec_CAF1 18934 119 + 1 UUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA-UUGGCGUGGAGUUUGUCUUUUCGAAUUGCCGCUAACAGAGCGCUGAGCUCCUCACGCGAACGAGAUCCACCGAC .....((((..(.(.(((((((((((((((((((((((((((((.-...)))....))))))....)))))))))))))....((((.....)))).....)))))))).)..))))... ( -47.80) >DroSim_CAF1 18998 119 + 1 UUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA-UUGGCGUGGAGUUUGUCUUUUUGAAUUGCCGCUAACAGAGCGCUGAGCUCCUCACGCGAACGAGAUCCACCGAC .....((((..(.(.(((((((((((((((((..((((((((((.-...)))....))))))....)..))))))))))....((((.....)))).....)))))))).)..))))... ( -45.60) >DroEre_CAF1 19262 119 + 1 UUUGAGGUGUUUGUGCGUUUGCAGCGGCGACUUGAACGAACCGUA-CUGGCGUGGAGUUUAUGUUUUCGAAUUGCCGCCAACGGAGCUCUGAGCUCCUCACGCGAACGAGUUCCACCGAC .....((((......(((((((.(((((((.(((((.((((..((-(....)))..)))).....))))).)))))))....(((((.....)))))....))))))).....))))... ( -42.60) >DroYak_CAF1 19406 120 + 1 UUUGAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGUACCGUUUUUGGCGUGGAGUUUAUCUUUUCGAAUUGCCGCUAACGGAGCGCUGAGCUCCACACGCGAACGAGAUCCACCGAC .....((((..(.(.(((((((((((((((((((((((..(((....)))..))(((.....)))))))))))))))))...(((((.....)))))....)))))))).)..))))... ( -52.10) >consensus UUUAAGGUGUUUGUGCGUUUGCAGCGGCGAUUUGAACGAACCGUA_UUGGCGUGGAGUUUGUCUUUUCGAAUUGCCGCUAACAGAGCGCUGAGCUCCUCACGCGAACGAGAUCCACCGAC .....((((......(((((((((((((((((((((((..(((....)))..))(((.....)))))))))))))))))....((((.....)))).....))))))).....))))... (-42.78 = -43.22 + 0.44)

| Location | 7,496,190 – 7,496,309 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -33.45 |
| Energy contribution | -33.09 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7496190 119 - 22407834 GUCGGUGGAUCUCGUUCGCGUGAGGAGCUCAGCGCUCUGUUAGCGGCAAUUCGAAAAGCCAAACUCCACGCCAA-UACGGUUCGUUCAAAUCGCCGCUGCAAACGCACAAACACCUUAAA ...((((((......))((((..(((((.....)))))((.((((((.(((.(((.((((..............-...))))..))).))).))))))))..)))).....))))..... ( -35.13) >DroSec_CAF1 18934 119 - 1 GUCGGUGGAUCUCGUUCGCGUGAGGAGCUCAGCGCUCUGUUAGCGGCAAUUCGAAAAGACAAACUCCACGCCAA-UACGGUUCGUUCAAAUCGCCGCUGCAAACGCACAAACACCUUAAA ...((((((......))((((..(((((.....)))))((.((((((.(((.(((..(((..............-....)))..))).))).))))))))..)))).....))))..... ( -34.07) >DroSim_CAF1 18998 119 - 1 GUCGGUGGAUCUCGUUCGCGUGAGGAGCUCAGCGCUCUGUUAGCGGCAAUUCAAAAAGACAAACUCCACGCCAA-UACGGUUCGUUCAAAUCGCCGCUGCAAACGCACAAACACCUUAAA ...((((((......))((((..(((((.....)))))((.((((((...((.....))..........(((..-...)))...........))))))))..)))).....))))..... ( -31.80) >DroEre_CAF1 19262 119 - 1 GUCGGUGGAACUCGUUCGCGUGAGGAGCUCAGAGCUCCGUUGGCGGCAAUUCGAAAACAUAAACUCCACGCCAG-UACGGUUCGUUCAAGUCGCCGCUGCAAACGCACAAACACCUCAAA ...(((((((....)))((((..(((((.....)))))...((((((.(((.(((......((((..((....)-)..))))..))).))).))))))....)))).....))))..... ( -37.00) >DroYak_CAF1 19406 120 - 1 GUCGGUGGAUCUCGUUCGCGUGUGGAGCUCAGCGCUCCGUUAGCGGCAAUUCGAAAAGAUAAACUCCACGCCAAAAACGGUACGUUCAAAUCGCCGCUGCAAACGCACAAACACCUCAAA ...((((((......))((((..(((((.....)))))((.((((((.(((.(((..((.....))...(((......)))...))).))).))))))))..)))).....))))..... ( -36.50) >consensus GUCGGUGGAUCUCGUUCGCGUGAGGAGCUCAGCGCUCUGUUAGCGGCAAUUCGAAAAGACAAACUCCACGCCAA_UACGGUUCGUUCAAAUCGCCGCUGCAAACGCACAAACACCUUAAA ...((((((......))((((..(((((.....)))))((.((((((.(((.(((..............(((......)))...))).))).))))))))..)))).....))))..... (-33.45 = -33.09 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:33 2006