
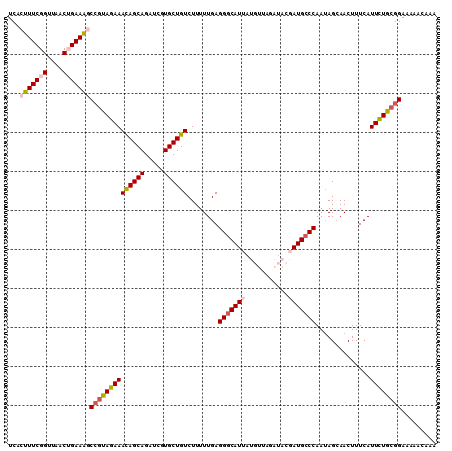
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,489,042 – 7,489,266 |
| Length | 224 |
| Max. P | 0.999365 |

| Location | 7,489,042 – 7,489,152 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -27.28 |
| Energy contribution | -28.58 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7489042 110 + 22407834 UCACUUUCGGUUAACUGAAAGCCGUAGAAACAGCAGAUCGUGCUGUCUUUUUGAGGGCAUUAUGUUAGAUACGAUGACCAAUAGAAACUUUCAUUCUGCGGAAAAACAAA ...(((((((....)))))))((((((((((((((.....)))))).((((((..(((((..(((...)))..))).))..))))))......))))))))......... ( -31.70) >DroSec_CAF1 11792 110 + 1 UCACUUUCGGUUAACAGAAAGCAGCAGAAACAGCAGAUCGUGCUGUCUUUUUGAGGGCAUUAUGUUAGAUACGAUGCCCAAUAGCAACUUUCAUUCUGCGGAAAAACAAA ...(((((........)))))..((((((((((((.....))))))........((((((..(((...)))..))))))..............))))))........... ( -30.60) >DroSim_CAF1 11916 110 + 1 UCACUUUCGGUGAACAGAAAGCCGUAGAAAUAGCAGAUCGUGCUGUCUUUUUGAGGGCAUUAUGUUAGAUACGAUGCCCAAUAGCAACUUUCAUUCUGCGGAAAAACAAA ...(((((.(....).)))))((((((((((((((.....))))))........((((((..(((...)))..))))))..............))))))))......... ( -32.60) >DroEre_CAF1 11774 104 + 1 UCACUUUCGGUUAACUGAAGCCCGUAGAAACAGCAGAUCGUGCUGUCUUUUUGAGGGCAU-----UAGAAACGAUGCCCAAAAGCAACUUUCAUUCUGGGGA-AAACAAA ....((((((....))))))(((.(((((((((((.....))))))........((((((-----(......)))))))..............)))))))).-....... ( -30.10) >DroYak_CAF1 12085 105 + 1 UCACUUUCGGUAAACUGAAGUCCGCAGAAACAGCAGAUCGUGCUGUCUUUUUGAGGGCAU-----UAGAAGCGAUGCCCAAUAGCAACUUUCAUUUUGCGGAAAAACAAA ....((((((....))))))(((((((((((((((.....))))))........((((((-----(......)))))))..............)))))))))........ ( -34.50) >consensus UCACUUUCGGUUAACUGAAAGCCGUAGAAACAGCAGAUCGUGCUGUCUUUUUGAGGGCAUUAUGUUAGAUACGAUGCCCAAUAGCAACUUUCAUUCUGCGGAAAAACAAA ...(((((((....)))))))((((((((((((((.....))))))........(((((((...........)))))))..............))))))))......... (-27.28 = -28.58 + 1.30)


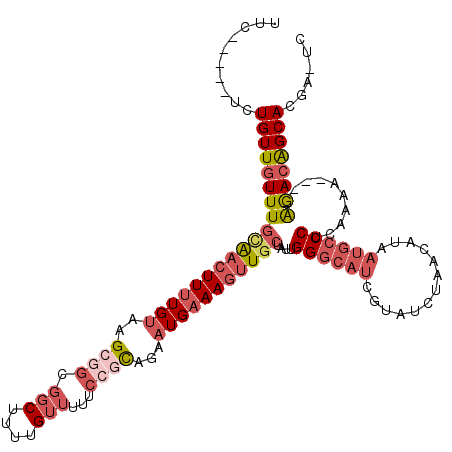
| Location | 7,489,077 – 7,489,187 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -20.48 |
| Energy contribution | -22.72 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7489077 110 - 22407834 UUC-----UCUGUUGUUUGCAACUUUUGUAAGCGGCGGCUUUUGUUUUUCCGCAGAAUGAAAGUUUCUAUUGGUCAUCGUAUCUAACAUAAUGCCCUCAAAA----AGACAGCACGA-UC ...-----(((((((((((.((((((..(..((((.(((....)))...))))...)..)))))).)....((.(((..(.......)..))).))......----)))))))).))-.. ( -21.70) >DroSec_CAF1 11827 110 - 1 UUC-----UCUGUUGUUUGCAACUUUUGUAAGCGGCGGCUUUUGUUUUUCCGCAGAAUGAAAGUUGCUAUUGGGCAUCGUAUCUAACAUAAUGCCCUCAAAA----AGACAGCACGA-UC ...-----((((((((((((((((((..(..((((.(((....)))...))))...)..))))))))....((((((..(.......)..))))))......----)))))))).))-.. ( -35.40) >DroSim_CAF1 11951 110 - 1 UUC-----UCUGUUGUUUGCAACUUUUGUAAGCGGCGGCUUUUGUUUUUCCGCAGAAUGAAAGUUGCUAUUGGGCAUCGUAUCUAACAUAAUGCCCUCAAAA----AGACAGCACGA-UC ...-----((((((((((((((((((..(..((((.(((....)))...))))...)..))))))))....((((((..(.......)..))))))......----)))))))).))-.. ( -35.40) >DroEre_CAF1 11809 104 - 1 UUC-----UCUGUUGUUUGCAACUUUUGUAAGCAUAGGCUUUUGUUU-UCCCCAGAAUGAAAGUUGCUUUUGGGCAUCGUUUCUA-----AUGCCCUCAAAA----AGACAGCACGA-UC ...-----((((((((((((((((((..(.(((....)))((((...-....)))))..))))))))....((((((........-----))))))......----)))))))).))-.. ( -30.10) >DroYak_CAF1 12120 105 - 1 UUC-----UCUGUUGUUUGCAACUUUUGUAAGCGGAGGCUUUUGUUUUUCCGCAAAAUGAAAGUUGCUAUUGGGCAUCGCUUCUA-----AUGCCCUCAAAA----AGACAGCACGA-UC ...-----((((((((((((((((((..(..(((((((........)))))))...)..))))))))....((((((........-----))))))......----)))))))).))-.. ( -37.60) >DroPer_CAF1 15229 104 - 1 UUCCUAGUUCUGUUCUUUGUG--UUGUGCAAGC-----CUUUUGUUUU--CUUUGAAUGAAAGUUGAAAUUGGGACUC-------CCCUACUGCCCUCAAACUAGGGAAUGGCACAAAGC ..............(((((((--(..(.(((..-----((((..(((.--....)))..))))))).....(((....-------))).....((((......)))).)..)))))))). ( -24.40) >consensus UUC_____UCUGUUGUUUGCAACUUUUGUAAGCGGCGGCUUUUGUUUUUCCGCAGAAUGAAAGUUGCUAUUGGGCAUCGUAUCUAACAUAAUGCCCUCAAAA____AGACAGCACGA_UC ..........(((((((((((((((((((..((((.(((....)))...))))...)))))))))))....((((((.............))))))..........))))))))...... (-20.48 = -22.72 + 2.24)

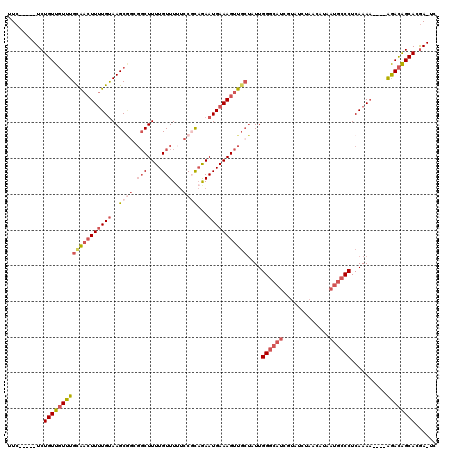
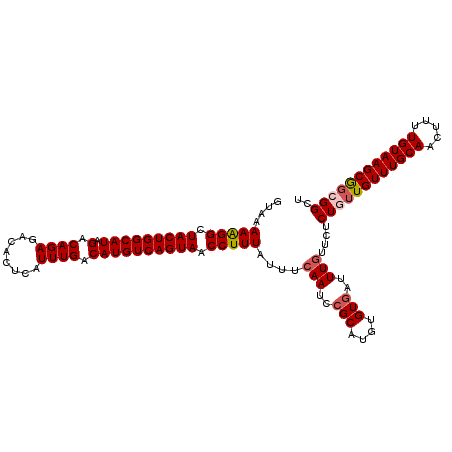
| Location | 7,489,152 – 7,489,266 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7489152 114 - 22407834 GUAAAAAGGCUACUGGCAUAGACAGAGACACUCAUUUGACAUGUCAGUAACCUUUAUUUCAAUCCGCAUGUGUGAUUUGUUCUCUGUUGUUUGCAACUUUUGUAAGCGGCGGCU ....(((((.(((((((((.(.((((........)))).)))))))))).)))))....(((..(((....)))..)))....((((((((((((.....)))))))))))).. ( -34.30) >DroSec_CAF1 11902 114 - 1 GUAAAAAGGCUACUGGCAUAGACAGAGACACUCAUUUGACAUGUCAGUAACCUUUAUUUCAAUCCGCAUGUGUGAUUUGUUCUCUGUUGUUUGCAACUUUUGUAAGCGGCGGCU ....(((((.(((((((((.(.((((........)))).)))))))))).)))))....(((..(((....)))..)))....((((((((((((.....)))))))))))).. ( -34.30) >DroSim_CAF1 12026 114 - 1 GUAAAAAGGCUACUGGCAUAGACAGAGACACUCAUUUGACAUGUCAGUAACCUUUAUUUGAAUUCGCAUGUGUGAUUUGUUCUCUGUUGUUUGCAACUUUUGUAAGCGGCGGCU ....(((((.(((((((((.(.((((........)))).)))))))))).)))))....((((((((....))))...)))).((((((((((((.....)))))))))))).. ( -35.40) >DroEre_CAF1 11878 114 - 1 GUAAAAAGGCUACUGGCAUAGACAGAGAGACUCAUUUGACAUGUCAGUAACCUUUAUUUCAAUCCGCAUGUGUGAUUUGUUCUCUGUUGUUUGCAACUUUUGUAAGCAUAGGCU ....(((((.(((((((((.(.((((........)))).)))))))))).)))))....(((..(((....)))..)))...(((((.(((((((.....)))))))))))).. ( -30.30) >DroYak_CAF1 12190 114 - 1 GUAAAAGGGCUACUGGCAUAGACAGAGAUACUCAUUUGACAUGUCAGUAACCUUUAUUUCAAUCCGCAUGUGUCAUUUGUUCUCUGUUGUUUGCAACUUUUGUAAGCGGAGGCU .......((((.((((((..((((((((....((..(((((((((....................).))))))))..)).))))))))...))).((....))...))).)))) ( -28.85) >consensus GUAAAAAGGCUACUGGCAUAGACAGAGACACUCAUUUGACAUGUCAGUAACCUUUAUUUCAAUCCGCAUGUGUGAUUUGUUCUCUGUUGUUUGCAACUUUUGUAAGCGGCGGCU ....(((((.(((((((((.(.((((........)))).)))))))))).)))))....(((..(((....)))..)))....((((((((((((.....)))))))))))).. (-30.30 = -30.98 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:24 2006