
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,467,462 – 7,467,602 |
| Length | 140 |
| Max. P | 0.948945 |

| Location | 7,467,462 – 7,467,562 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -23.88 |
| Energy contribution | -25.72 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7467462 100 - 22407834 ACCAAAAGAGGACAGCUGCUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUCACGGCAUUGAAAUUGUUAAACCGUUGACCUGUCAAAGCCGACUU .(((...((((((........)))))).(((((....)))))...))).........((((.((((....(((((....)))))...)))).)))).... ( -33.20) >DroSec_CAF1 62954 100 - 1 ACCAAAAGAGGACAGCUGUUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUCACGGCAUUGAAAUUGUUAAACCGUUGACCUGUCCAAGCCGACUU .(((...((((((........)))))).(((((....)))))...))).........((((.(((.....(((((....))))).....))))))).... ( -30.00) >DroSim_CAF1 65422 100 - 1 ACCAAAAGAGGACAGCUGUUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUUACGGCAUUGAAAUUGUUAAACCGUUGACCUGUCAAAGCCGACUU .(((...((((((........)))))).(((((....)))))...))).........((((.((((....(((((....)))))...)))).)))).... ( -33.20) >DroEre_CAF1 66624 95 - 1 GCCAAAAGAGGACAGCUGUGAGUCCUCCUCCGCAAUUGCG----AUGGGAAAAUUCACGGCAUUGAAAUGGUUAAGCCGUUGACCUGCCA-AGCCGACUU .((......)).(.(((((((((..((((.(((....)))----..))))..))))))((((....(((((.....)))))....)))).-))).).... ( -30.10) >DroYak_CAF1 66080 96 - 1 ACCAAAAGCGGACAGCUGUGACUACUCCUCCGCAAUUGCG----AUGGGAAAAUGCACGGCAUUGAAAUUGUUAAAUCGUUGACCUGUCAAAGUCGACUU .....(((..(((.((((((.....((((.(((....)))----..)))).....)))))).((((....(((((....)))))...)))).)))..))) ( -25.40) >consensus ACCAAAAGAGGACAGCUGUUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUCACGGCAUUGAAAUUGUUAAACCGUUGACCUGUCAAAGCCGACUU .(((...((((((........)))))).(((((....)))))...))).........((((.((((....(((((....)))))...)))).)))).... (-23.88 = -25.72 + 1.84)

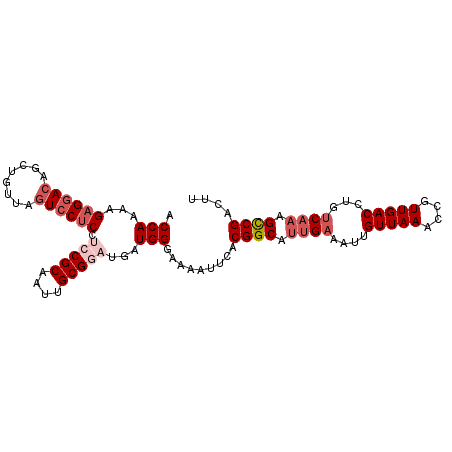
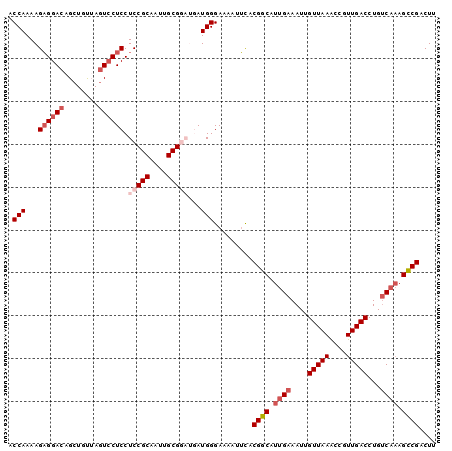
| Location | 7,467,483 – 7,467,602 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -23.05 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7467483 119 + 22407834 CGGUUUAACAAUUUCAAUGCCGUGAAUUUUCCCAUCAUCCGCAAUUGCGGAGGAGGACUAGCAGCUGUCCUCUUUUGGUCUGCAAUCUCCGAAAAUUCCACAGUAGAAAUCACCAGUCA .(((......(((((..(((.((((((((((......(((((....)))))(((((....((((....((......)).))))..)))))))))))).))).)))))))).)))..... ( -33.70) >DroSec_CAF1 62975 119 + 1 CGGUUUAACAAUUUCAAUGCCGUGAAUUUUCCCAUCAUCCGCAAUUGCGGAGGAGGACUAACAGCUGUCCUCUUUUGGUCUGCAAUCUCCGAAAAUUCCACAGUAGAAAUCUCCAGUCA .((.......(((((..(((.((((((((((......(((((....)))))(((((((........))))))).................))))))).))).))))))))..))..... ( -31.10) >DroSim_CAF1 65443 119 + 1 CGGUUUAACAAUUUCAAUGCCGUAAAUUUUCCCAUCAUCCGCAAUUGCGGAGGAGGACUAACAGCUGUCCUCUUUUGGUCUGCAAUCUCCGAAAAUUCCACAGUAGAAAUCUCCAGUCA .((.......(((((..(((.((.(((((((......(((((....)))))(((((((........))))))).................)))))))..)).))))))))..))..... ( -27.20) >DroEre_CAF1 66644 115 + 1 CGGCUUAACCAUUUCAAUGCCGUGAAUUUUCCCAU----CGCAAUUGCGGAGGAGGACUCACAGCUGUCCUCUUUUGGCCUGCAAUCUGCGAAAAUUCCACAGUAGAAAUCUCCAGUCA .((((.....(((((..(((.((((((((.....(----(((((((((((((((((((........)))))))).....))))))).)))))))))).))).))))))))....)))). ( -36.00) >DroYak_CAF1 66101 115 + 1 CGAUUUAACAAUUUCAAUGCCGUGCAUUUUCCCAU----CGCAAUUGCGGAGGAGUAGUCACAGCUGUCCGCUUUUGGUCUGCAAUCUCCGAAAAUUCCACAGUAGAAAUCUGCAGUCA .((((................(((.((((((....----.((....))(((((.(((((((.(((.....)))..))).))))..)))))))))))..))).((((....)))))))). ( -27.20) >consensus CGGUUUAACAAUUUCAAUGCCGUGAAUUUUCCCAUCAUCCGCAAUUGCGGAGGAGGACUAACAGCUGUCCUCUUUUGGUCUGCAAUCUCCGAAAAUUCCACAGUAGAAAUCUCCAGUCA .((((.....(((((..(((.((((((((((.........(((....(((((((((((........)))))))))))...))).......))))))).))).))))))))....)))). (-23.05 = -23.73 + 0.68)

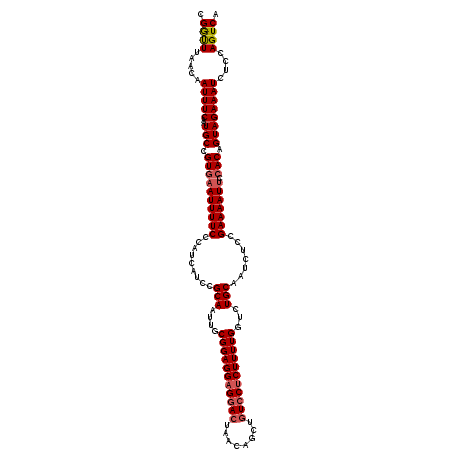
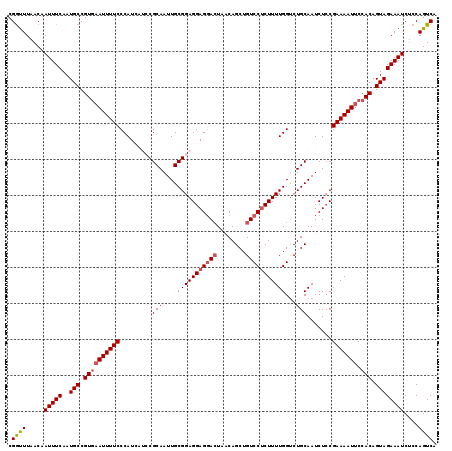
| Location | 7,467,483 – 7,467,602 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -28.28 |
| Energy contribution | -29.56 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7467483 119 - 22407834 UGACUGGUGAUUUCUACUGUGGAAUUUUCGGAGAUUGCAGACCAAAAGAGGACAGCUGCUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUCACGGCAUUGAAAUUGUUAAACCG ....(((..(((((..(((((((.((((((((((((((((.((......))....)))).))).))))(((((....))))).....))))).)))))))....)))))..)))..... ( -38.90) >DroSec_CAF1 62975 119 - 1 UGACUGGAGAUUUCUACUGUGGAAUUUUCGGAGAUUGCAGACCAAAAGAGGACAGCUGUUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUCACGGCAUUGAAAUUGUUAAACCG ((((....((((((..(((((((.((((((((((((((((.((......))....))).)))).))))(((((....))))).....))))).)))))))....))))))))))..... ( -34.50) >DroSim_CAF1 65443 119 - 1 UGACUGGAGAUUUCUACUGUGGAAUUUUCGGAGAUUGCAGACCAAAAGAGGACAGCUGUUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUUACGGCAUUGAAAUUGUUAAACCG ...(((((((((((......)))))))))))(((((.....(((...((((((........)))))).(((((....)))))...)))...))))).(((..((((.....)))).))) ( -33.70) >DroEre_CAF1 66644 115 - 1 UGACUGGAGAUUUCUACUGUGGAAUUUUCGCAGAUUGCAGGCCAAAAGAGGACAGCUGUGAGUCCUCCUCCGCAAUUGCG----AUGGGAAAAUUCACGGCAUUGAAAUGGUUAAGCCG .....(((((((((......)))))))))((.....)).(((............(((((((((..((((.(((....)))----..))))..))))))))).((((.....))))))). ( -35.00) >DroYak_CAF1 66101 115 - 1 UGACUGCAGAUUUCUACUGUGGAAUUUUCGGAGAUUGCAGACCAAAAGCGGACAGCUGUGACUACUCCUCCGCAAUUGCG----AUGGGAAAAUGCACGGCAUUGAAAUUGUUAAAUCG ...(((.(((((((......))))))).))).((((((((.((......))...((((((.....((((.(((....)))----..)))).....)))))).......))))..)))). ( -29.80) >consensus UGACUGGAGAUUUCUACUGUGGAAUUUUCGGAGAUUGCAGACCAAAAGAGGACAGCUGUUAGUCCUCCUCCGCAAUUGCGGAUGAUGGGAAAAUUCACGGCAUUGAAAUUGUUAAACCG ((((....((((((..(((((((.((((((((((((((((.((......))....)))).))).))))(((((....))))).....))))).)))))))....))))))))))..... (-28.28 = -29.56 + 1.28)

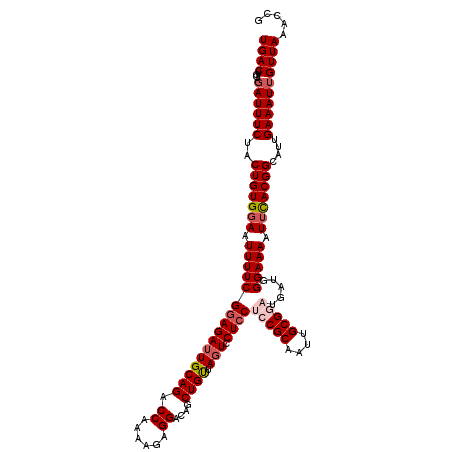

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:12 2006