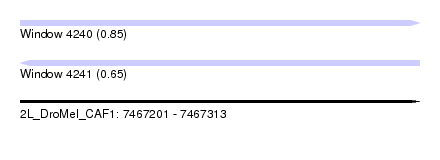
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,467,201 – 7,467,313 |
| Length | 112 |
| Max. P | 0.851148 |

| Location | 7,467,201 – 7,467,313 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -27.56 |
| Energy contribution | -27.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
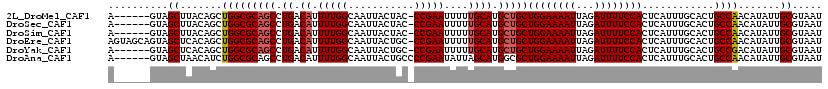
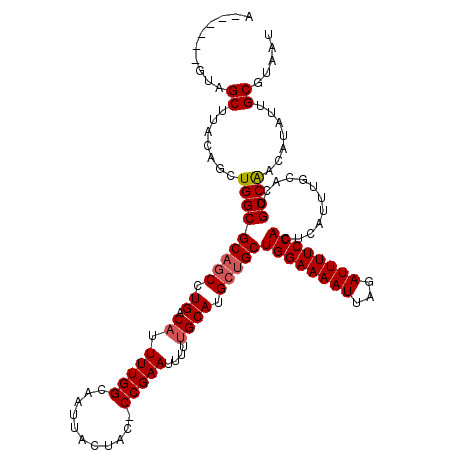
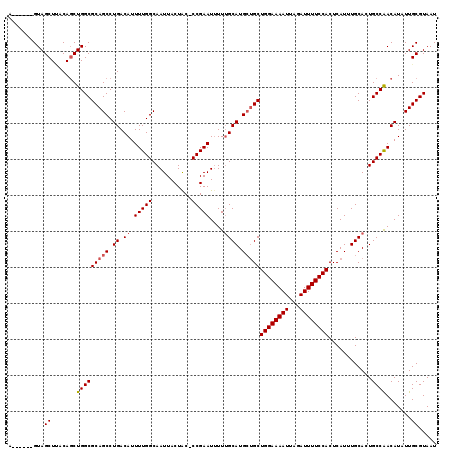
>2L_DroMel_CAF1 7467201 112 + 22407834 AUUACGCAAUAUGUUGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCAGCAUGCAAAAAUUCGG-GUAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGCUGUAAGCUAC------U .....((.....((((((............((((((((...))))))))(((((.((.(...(((..(-((((...)))))..))).))).))))))))))).....))...------. ( -31.50) >DroSec_CAF1 62693 112 + 1 AUUACGCAAUAUGUUGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCAGCAUGCAAAAAUUCGG-GUAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGCUGUAAGCUAC------U .....((.....((((((............((((((((...))))))))(((((.((.(...(((..(-((((...)))))..))).))).))))))))))).....))...------. ( -31.50) >DroSim_CAF1 65161 112 + 1 AUUACGCAAUAUGUUGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCAGCAUGCAAAAAUUCGG-GUAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGCUGUAAGCUAC------U .....((.....((((((............((((((((...))))))))(((((.((.(...(((..(-((((...)))))..))).))).))))))))))).....))...------. ( -31.50) >DroEre_CAF1 66357 118 + 1 AUUACGCAAUAUGUUGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCAGCAUGCAAAAAUUCGG-GCAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGCUGUGAGCUACUGCUACU ....((((....((((((............((((((((...))))))))(((((.((.(...(((..(-((((...)))))..))).))).)))))))))))))))(((....)))... ( -36.40) >DroYak_CAF1 65812 112 + 1 AUUACGCAAUAUGUCGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCAGCAUGCAAAAAUUCGG-GCAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGCUGUGAGCUAC------U .....((.(((.((.(((............((((((((...))))))))(((((.((.(...(((..(-((((...)))))..))).))).)))))))).)))))..))...------. ( -30.40) >DroAna_CAF1 63871 113 + 1 AUUACGCAAUAUGUUGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCGCCAUGCUAAUAUUCGGGGCAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGAUGUUAGCUAC------U .....(((...((....)).)))..(((.(((((((((...)))))))).)..)))((((((((..((.(((((...((.(.....).)).))))).))..))))))))...------. ( -34.00) >consensus AUUACGCAAUAUGUUGGCAGUGCAAAUGAGUGGAAAAUCUAAUUUUCCAGCAGCAUGCAAAAAUUCGG_GCAGUAAUUGCCAAAAUGUCAGGCUGCGCCAGCUGUAAGCUAC______U .....((.....((((((..((((..((..((((((((...)))))))).))...))))..........(((((...((.(.....).)).))))))))))).....)).......... (-27.56 = -27.67 + 0.11)

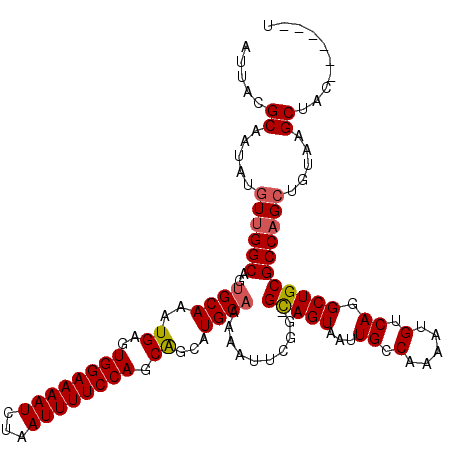
| Location | 7,467,201 – 7,467,313 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7467201 112 - 22407834 A------GUAGCUUACAGCUGGCGCAGCCUGACAUUUUGGCAAUUACUAC-CCGAAUUUUUGCAUGCUGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCAACAUAUUGCGUAAU .------(((((.....))(((((((((.((.((.(((((..........-)))))....)))).)))))((((((((...))))))))............))))......)))..... ( -27.00) >DroSec_CAF1 62693 112 - 1 A------GUAGCUUACAGCUGGCGCAGCCUGACAUUUUGGCAAUUACUAC-CCGAAUUUUUGCAUGCUGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCAACAUAUUGCGUAAU .------(((((.....))(((((((((.((.((.(((((..........-)))))....)))).)))))((((((((...))))))))............))))......)))..... ( -27.00) >DroSim_CAF1 65161 112 - 1 A------GUAGCUUACAGCUGGCGCAGCCUGACAUUUUGGCAAUUACUAC-CCGAAUUUUUGCAUGCUGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCAACAUAUUGCGUAAU .------(((((.....))(((((((((.((.((.(((((..........-)))))....)))).)))))((((((((...))))))))............))))......)))..... ( -27.00) >DroEre_CAF1 66357 118 - 1 AGUAGCAGUAGCUCACAGCUGGCGCAGCCUGACAUUUUGGCAAUUACUGC-CCGAAUUUUUGCAUGCUGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCAACAUAUUGCGUAAU ....((((((((.....))(((((((((.((..((((.((((.....)))-).)))).....)).)))))((((((((...))))))))............))))...))))))..... ( -35.50) >DroYak_CAF1 65812 112 - 1 A------GUAGCUCACAGCUGGCGCAGCCUGACAUUUUGGCAAUUACUGC-CCGAAUUUUUGCAUGCUGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCGACAUAUUGCGUAAU .------(((((.....))(((((((((.((..((((.((((.....)))-).)))).....)).)))))((((((((...))))))))............))))......)))..... ( -29.40) >DroAna_CAF1 63871 113 - 1 A------GUAGCUAACAUCUGGCGCAGCCUGACAUUUUGGCAAUUACUGCCCCGAAUAUUAGCAUGGCGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCAACAUAUUGCGUAAU .------((((((((.((.(((.((((((.........)))......))).))).)).))))).((((((((((((((...)))))))).......))...))))......)))..... ( -25.61) >consensus A______GUAGCUUACAGCUGGCGCAGCCUGACAUUUUGGCAAUUACUAC_CCGAAUUUUUGCAUGCUGCUGGAAAAUUAGAUUUUCCACUCAUUUGCACUGCCAACAUAUUGCGUAAU ..........((.......(((((((((.((.((.(((((...........)))))....)))).)))))((((((((...))))))))............)))).......))..... (-24.61 = -24.97 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:10 2006