
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,463,420 – 7,463,542 |
| Length | 122 |
| Max. P | 0.733656 |

| Location | 7,463,420 – 7,463,511 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
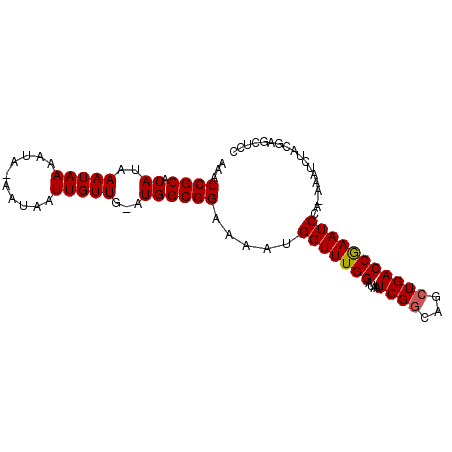
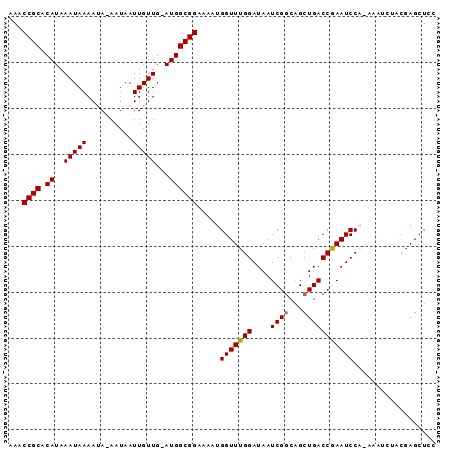
>2L_DroMel_CAF1 7463420 91 + 22407834 AAACCGCACAUAAAUAAAAGA-AAUAAUUGUUG-AUGGCGGAAAAUGGUUUGGAUAAUCGGCAGCUGACCAAAUCCAAAAAUCUACGAGCUCC ...((((.(((.(((((....-.....))))).-)))))))....((((((((....((((...))))))))).)))................ ( -17.50) >DroSec_CAF1 59035 91 + 1 AAACCGCACAUAAAUAAAAUA-AAUAAUUGUUG-AUGGCGGAAAAUGGUUUGGAUAAUCGGCAGCUGACCGAAUCCACAAAUCUGCGAGCUCC ...((((.(((.(((((....-.....))))).-))))))).....(((((((((..(((((....).)))))))))..)))).......... ( -20.00) >DroSim_CAF1 61062 91 + 1 AAACCGCACAUAAAUAAAAUA-AAUAAUUGUUG-AUGGCGGAAAAUGGUUUGGAUAAUCGGCAGCUGACCGAAUCCACAAAUCUACGAGCUCC ...((((.(((.(((((....-.....))))).-))))))).....(((((((((..(((((....).)))))))))..)))).......... ( -20.00) >DroEre_CAF1 62528 83 + 1 AAACCGCACAUAAAUAAAACA-AAUAAUUGUUG-AUGGCGGAAAAUGGUUUGGAUAAUCGGCUGCUGACCGAAUCC--------GCGAGCUCC ...((((.(((......((((-(....))))).-))))))).....(((((((((..(((((....).))))))))--------..))))).. ( -20.40) >DroYak_CAF1 61949 84 + 1 AAACCGC-CAUAAAUAAAAUAAAAUAAUUGUUGAAUGGCGGAAAAUGGUUUGGAUAAUCGCCAGCUGACCAAAUCC--------ACGAGCUGC ...((((-(((.(((((..........)))))..)))))))....((((((((.......)))...))))).....--------......... ( -18.80) >consensus AAACCGCACAUAAAUAAAAUA_AAUAAUUGUUG_AUGGCGGAAAAUGGUUUGGAUAAUCGGCAGCUGACCGAAUCCA_AAAUCUACGAGCUCC ...((((.((..(((((..........)))))...)))))).....(((((((....((((...))))))))))).................. (-14.04 = -14.00 + -0.04)

| Location | 7,463,438 – 7,463,542 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -16.07 |
| Energy contribution | -17.23 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
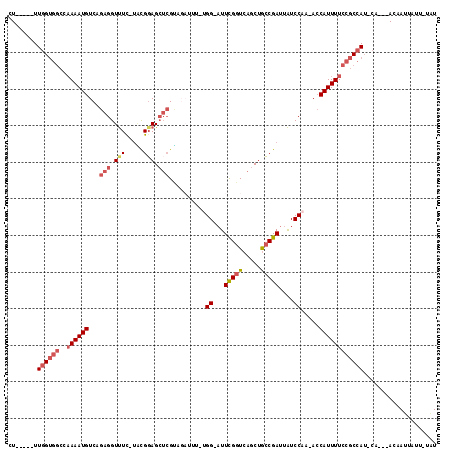
>2L_DroMel_CAF1 7463438 104 - 22407834 CU-----UUGGUGGCGAAAAUGUCAGAGGUUUC-UACGGAGCUCGUAGAUUUUUGG-AUUUGGUCAGCUGCCGAUUAUCCAA-ACCAUUUUCCGCCAU-CA---ACAAUUAUU-UCU ..-----(((((((((((((((.........((-((((.....))))))..(((((-(((((((.....)))))..))))))-).)))))).))))))-))---)........-... ( -34.40) >DroSec_CAF1 59053 104 - 1 CU-----UUGGUGGCCAAAAUGUCAGAGGUUUC-UACGGAGCUCGCAGAUUUGUGG-AUUCGGUCAGCUGCCGAUUAUCCAA-ACCAUUUUCCGCCAU-CA---ACAAUUAUU-UAU ..-----((((((((.((((((...(((..(((-....)))))).........(((-(((((((.....)))))..))))).-..))))))..)))))-))---)........-... ( -29.20) >DroSim_CAF1 61080 104 - 1 CU-----UUGGUGGCCAAAAUGUCAGAGGUUUC-UACGGAGCUCGUAGAUUUGUGG-AUUCGGUCAGCUGCCGAUUAUCCAA-ACCAUUUUCCGCCAU-CA---ACAAUUAUU-UAU ..-----((((((((.((((((.........((-((((.....))))))....(((-(((((((.....)))))..))))).-..))))))..)))))-))---)........-... ( -30.70) >DroEre_CAF1 62546 96 - 1 CU-----UUGGUGGCGAAAAUGUCAGAGGUUUC-UGCGGAGCUCGC--------GG-AUUCGGUCAGCAGCCGAUUAUCCAA-ACCAUUUUCCGCCAU-CA---ACAAUUAUU-UGU ..-----(((((((((((((((...(((.((..-....)).)))..--------((-(((((((.....)))))..))))..-..)))))).))))))-))---)........-... ( -32.20) >DroYak_CAF1 61966 98 - 1 UU-----UUGGUGGCGAAAAUGUCAGAGGUUUC-UGCGCAGCUCGU--------GG-AUUUGGUCAGCUGGCGAUUAUCCAA-ACCAUUUUCCGCCAUUCA---ACAAUUAUUUUAU ..-----(((((((((((((((...(((.(...-.....).))).(--------((-((.(.(((....))).)..))))).-..)))))).))))).)))---)............ ( -24.10) >DroAna_CAF1 60217 112 - 1 UUUUUUUUUUGCCUCCCAAAUGUCAGAGGUCUUGCUUGGAG---GAUGAUCUUUGGUCUUUGGUCAGCUACCGAUUGUCCAAAAGCAUUUUCCACCAA-CACCGACCAUAUUU-UAU .................((((((....(((.(((..(((((---((((...(((((.(.(((((.....)))))..).)))))..))))))))).)))-.)))....))))))-... ( -27.10) >consensus CU_____UUGGUGGCCAAAAUGUCAGAGGUUUC_UACGGAGCUCGUAGAUUU_UGG_AUUCGGUCAGCUGCCGAUUAUCCAA_ACCAUUUUCCGCCAU_CA___ACAAUUAUU_UAU ........((((((..((((((...(((.(((.....))).)))..........((...(((((.....)))))....)).....)))))))))))).................... (-16.07 = -17.23 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:08 2006