| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,459,798 – 7,459,888 |
| Length | 90 |
| Max. P | 0.500000 |

| Location | 7,459,798 – 7,459,888 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -11.92 |
| Energy contribution | -14.54 |
| Covariance contribution | 2.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
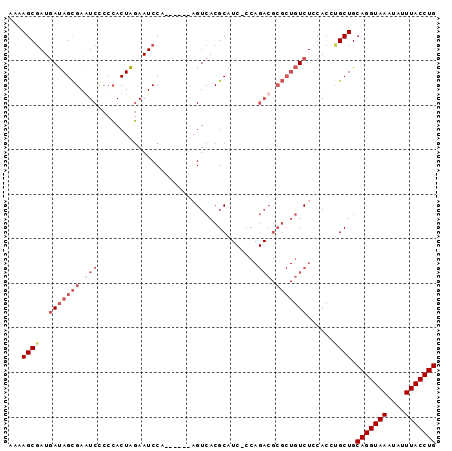
>2L_DroMel_CAF1 7459798 90 + 22407834 AAAAGCGAUGAUAGCGAAUCCCCCACUAGAAUCCA------AGUCACGCAUCCCCAGACGCGCUGUCUUCACUCGCUGCAGGUAAAUAUUUACCUG ...(((((.((((((((.((........)).))..------......((.((....)).)))))))).....))))).(((((((....))))))) ( -23.00) >DroSec_CAF1 55906 89 + 1 AAAAGCGAUGAUAGCGAGUCCCCCACUAGAAUCCA------AGUCACGCAUC-CCAGACGCGCUGUCUCCACCUGCUGCAGGUAAUUAUUUACCUG ...((((..(((((((.(((...............------...........-...))).)))))))......)))).(((((((....))))))) ( -21.45) >DroSim_CAF1 57448 89 + 1 AAAAGCGAUGAUAGCGAGUCCCCGACUACAAUCCA------AGUCACGCAUC-CCAGACGCGCUGUCUCCACCUGCUGCAGGUAAAUAUUUACCUG ...((((..(((((((.(((...((((........------))))..(....-.).))).)))))))......)))).(((((((....))))))) ( -24.40) >DroYak_CAF1 57539 81 + 1 AAAAGCAAUUACAGUGAAUCUCCCACUGGAAUCCCCUUUCCCGUCACGC---------------AUCUCCACCAGCUGCAGGUAAAUAUUUACCUG ...(((.....(((((.......)))))(((......))).........---------------..........))).(((((((....))))))) ( -15.80) >consensus AAAAGCGAUGAUAGCGAAUCCCCCACUAGAAUCCA______AGUCACGCAUC_CCAGACGCGCUGUCUCCACCUGCUGCAGGUAAAUAUUUACCUG ...((((..(((((((.(((....................................))).)))))))......)))).(((((((....))))))) (-11.92 = -14.54 + 2.62)

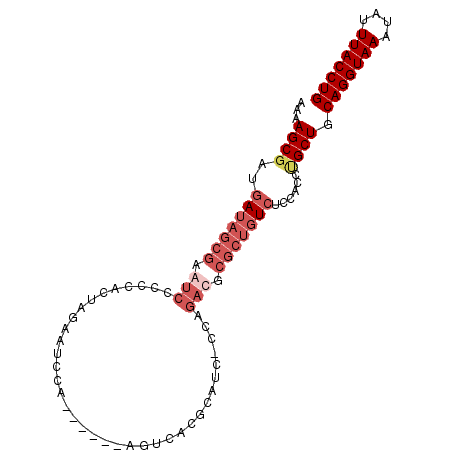
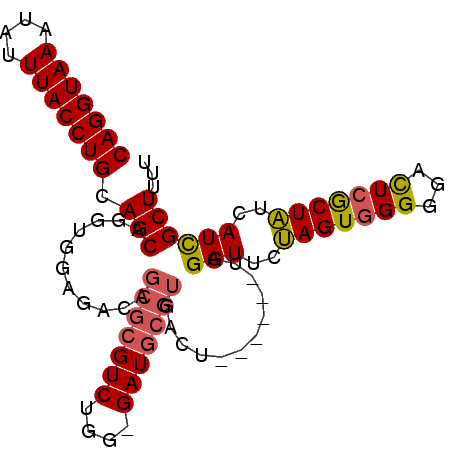
| Location | 7,459,798 – 7,459,888 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7459798 90 - 22407834 CAGGUAAAUAUUUACCUGCAGCGAGUGAAGACAGCGCGUCUGGGGAUGCGUGACU------UGGAUUCUAGUGGGGGAUUCGCUAUCAUCGCUUUU (((((((....)))))))....((((((.((.((((((((....)))))......------.(((((((.....)))))))))).)).)))))).. ( -31.40) >DroSec_CAF1 55906 89 - 1 CAGGUAAAUAAUUACCUGCAGCAGGUGGAGACAGCGCGUCUGG-GAUGCGUGACU------UGGAUUCUAGUGGGGGACUCGCUAUCAUCGCUUUU (((((((....)))))))...((((((....).(((((((...-))))))).)))------))(((..((((((....).)))))..)))...... ( -30.60) >DroSim_CAF1 57448 89 - 1 CAGGUAAAUAUUUACCUGCAGCAGGUGGAGACAGCGCGUCUGG-GAUGCGUGACU------UGGAUUGUAGUCGGGGACUCGCUAUCAUCGCUUUU (((((((....)))))))...((((((....).(((((((...-))))))).)))------))(((.(((((.(....)..))))).)))...... ( -29.20) >DroYak_CAF1 57539 81 - 1 CAGGUAAAUAUUUACCUGCAGCUGGUGGAGAU---------------GCGUGACGGGAAAGGGGAUUCCAGUGGGAGAUUCACUGUAAUUGCUUUU ((.(((....((((((.......))))))..)---------------)).)).....(((((.((((.((((((.....)))))).)))).))))) ( -20.80) >consensus CAGGUAAAUAUUUACCUGCAGCAGGUGGAGACAGCGCGUCUGG_GAUGCGUGACU______UGGAUUCUAGUGGGGGACUCGCUAUCAUCGCUUUU (((((((....))))))).(((...........(((((((....)))))))............(((..(((((((...)))))))..))))))... (-21.14 = -21.57 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:06 2006