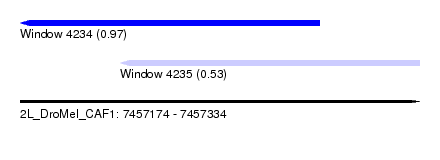
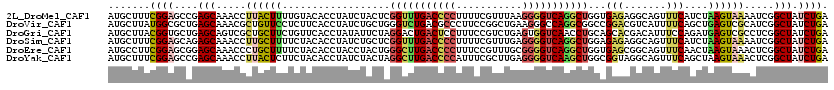
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,457,174 – 7,457,334 |
| Length | 160 |
| Max. P | 0.972442 |

| Location | 7,457,174 – 7,457,294 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.70 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
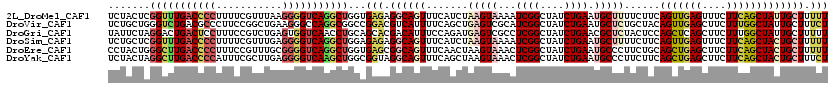
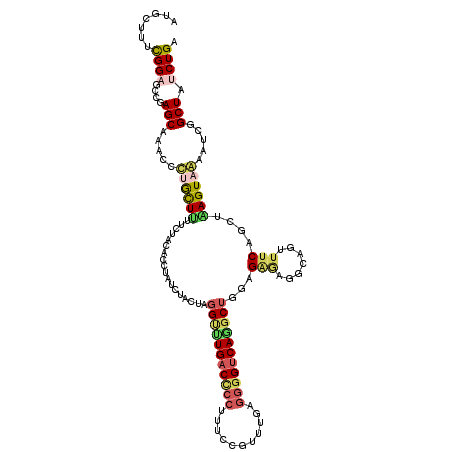
>2L_DroMel_CAF1 7457174 120 - 22407834 UCUACUCGGUUUGACCCCUUUUCGUUUAAGGGGUCAGGCUGGUGAGAGGCAGUUUCAUCUAAGUAAAAUCGGCUAUCUGAAUGCUUUUCUUCAGUUGAGUUUCUUCAGCUAUUGCUUUUU ..(((.((((((((((((((.......)))))))))))))))))(((((((((.......(((((...((((....)))).)))))......(((((((....)))))))))))))))). ( -40.50) >DroVir_CAF1 63369 120 - 1 UCUGCUGGGUCUGACGCCCUUCCGGCUGAAGGGCCAGGCGGCCGGACGUCAUUUUCAGCUGAGUCGCAUCGGCUAUCUGAAUGCUCUGCUACAGUUGAGCUUCUUUGGCUAUUGCUUUCU ((.((((((..(((((.((..(((.(((......))).)))..)).)))))..)))))).))...(((..(((((...(((.((((.((....)).)))))))..)))))..)))..... ( -48.70) >DroGri_CAF1 57749 120 - 1 UAUUCUAGGACUGACUCCUUUCCGUCUGAGUGGUCAACCUGCAGCACGACAUUUCCAGAUGAGUCGCCUCGGCUAUCUGAACGCUCUACUCCAGCUCAGCUUCUUUGGCUAUUGCUUUUU .......(((..(((........))).((((((((............))).....(((((.(((((...))))))))))..)))))...)))(((..((((.....))))...))).... ( -27.90) >DroSim_CAF1 54866 120 - 1 UCUGCUCGGUUUGACCCCUUUUCGUUUGAGGGGUCAGGCUGGAGAGAGGCAGUUUCAUCUAAGUAAAAUCGGCUAUCUGAAUGCUUUUCUUCAGUUGAGUUUCUUCAGCUACUGCUUUUU ...((..(((((((((((((.......)))))))))))))(((((((((((..((((....(((.......)))...)))))))))))))))(((((((....)))))))...))..... ( -43.30) >DroEre_CAF1 56251 120 - 1 CCUACUGGGCUUGACCCCUUUCCGUUUGCGGGGUCAGGCUGGUGAGCGGCAGUUUCAACUAAGUAAACUCGGCUAUCUGAAUGCCCUUCUGCAGCUGAGCUUCUUCAGCUACUGCUUUUU ..(((..(.(((((((((...........))))))))))..)))...((((((.........(((.....(((.........)))....)))(((((((....))))))))))))).... ( -40.40) >DroYak_CAF1 54919 120 - 1 UCUACUAGGCUUGACCCCAUUUCGCUUGAGGGGUCAAGCUGGCGGUAGGCAGUUUCAGCUAAGUAAACUCGGCUAUCUGAAUGCCCUUCUUCAGCUGAGCUUCUUCAGCUACUGCUUUCU .(((((.(((((((((((.((......)))))))))))))...)))))(((((...(((((((.((.(((((((....(((.....)))...))))))).))))).)))))))))..... ( -48.30) >consensus UCUACUAGGUUUGACCCCUUUCCGUUUGAGGGGUCAGGCUGGAGAGAGGCAGUUUCAGCUAAGUAAAAUCGGCUAUCUGAAUGCUCUUCUUCAGCUGAGCUUCUUCAGCUACUGCUUUUU .......(((((((((((...........)))))))))))...(((.((((((.......(((((...((((....)))).)))))......(((((((....))))))))))))).))) (-24.25 = -25.70 + 1.45)
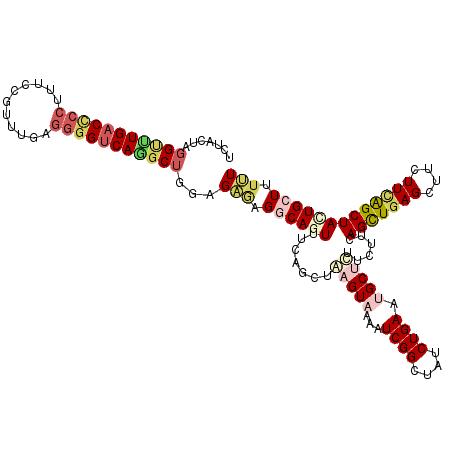
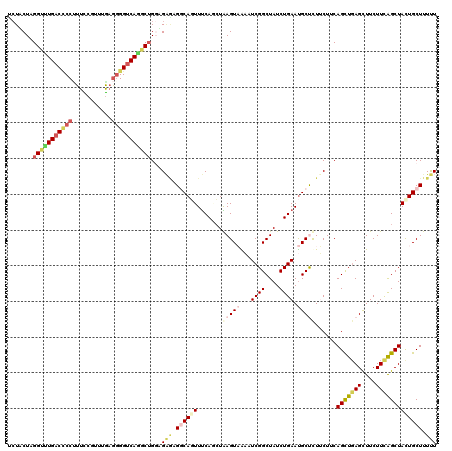
| Location | 7,457,214 – 7,457,334 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -20.07 |
| Energy contribution | -20.63 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7457214 120 - 22407834 AUGCUUUCGGAGCCGAGCAAACCUUACUUUUGUACACCUAUCUACUCGGUUUGACCCCUUUUCGUUUAAGGGGUCAGGCUGGUGAGAGGCAGUUUCAUCUAAGUAAAAUCGGCUAUCUGA ......((((((((((.......((((((.((.((.(((.((.((.((((((((((((((.......)))))))))))))))))).)))..))..))...))))))..)))))..))))) ( -43.60) >DroVir_CAF1 63409 120 - 1 AUGCUUAUGGCGCUGAGCAAACGCUGUUCCUCUUCACCUAUCUGCUGGGUCUGACGCCCUUCCGGCUGAAGGGCCAGGCGGCCGGACGUCAUUUUCAGCUGAGUCGCAUCGGCUAUCUGA ..((.....))(((((((....))(((..(((...........((((((..(((((.((..(((.(((......))).)))..)).)))))..)))))).)))..))))))))....... ( -40.00) >DroGri_CAF1 57789 120 - 1 AUGCUUACGGUGCUGAGCAGUCGCUGCUUCUGUUCACCUAUAUUCUAGGACUGACUCCUUUCCGUCUGAGUGGUCAACCUGCAGCACGACAUUUCCAGAUGAGUCGCCUCGGCUAUCUGA .((((((......))))))((((((((.........((((.....))))..((((..(((.......)))..))))....)))))..))).....(((((.(((((...)))))))))). ( -36.00) >DroSim_CAF1 54906 120 - 1 AUGCUUUCGGAGCAGAGCAAACCUUGCUUUUCUACACCUAUCUGCUCGGUUUGACCCCUUUUCGUUUGAGGGGUCAGGCUGGAGAGAGGCAGUUUCAUCUAAGUAAAAUCGGCUAUCUGA .(((((..(((((((((((.....))))))......(((.(((.(.((((((((((((((.......)))))))))))))))))).)))..)))))....)))))...((((....)))) ( -41.20) >DroEre_CAF1 56291 120 - 1 AUGCCUUCGGAGCGGAGCAAACCCUGCUUUUCUACACCUACCUACUGGGCUUGACCCCUUUCCGUUUGCGGGGUCAGGCUGGUGAGCGGCAGUUUCAACUAAGUAAACUCGGCUAUCUGA ......((((((..(((((.....)))))..)..((((..((....))((((((((((...........)))))))))).))))(((.(.((((((......).)))))).))).))))) ( -41.60) >DroYak_CAF1 54959 120 - 1 AUGCUUUCGGAGCCGAGCAAACCUUACUCUUCUACACCUAUCUACUAGGCUUGACCCCAUUUCGCUUGAGGGGUCAAGCUGGCGGUAGGCAGUUUCAGCUAAGUAAACUCGGCUAUCUGA ......(((((((((((......(((((...((....((..(((((.(((((((((((.((......)))))))))))))...)))))..))....))...))))).))))))..))))) ( -43.30) >consensus AUGCUUUCGGAGCCGAGCAAACCCUGCUUUUCUACACCUAUCUACUAGGUUUGACCCCUUUCCGUUUGAGGGGUCAGGCUGGAGAGAGGCAGUUUCAGCUAAGUAAAAUCGGCUAUCUGA .......((((....(((.....((((((..................(((((((((((...........)))))))))))...(((.......)))....)))))).....))).)))). (-20.07 = -20.63 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:04 2006