| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,448,644 – 7,448,737 |
| Length | 93 |
| Max. P | 0.500000 |

| Location | 7,448,644 – 7,448,737 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7448644 93 - 22407834 AUUCCUA--------C--UCGUAUUCAUGUGCAAUA--ACGCAAAGUGUCAAAGACCCAA-C---CAUAGCUCACUGCGGGGUCAAUUGAUAGACUCAAUUAUGACGAU .......--------.--.........((((.....--.))))...(((((..(((((..-.---....((.....)).)))))((((((.....)))))).))))).. ( -19.30) >DroVir_CAF1 51162 94 - 1 UUUUGUA--------CGGGCAU-UUGGUGCGGGAGA--ACCGUAGGUGUCAAAGACCCAA-C---CAUAGCUCACAUCGGGGUCAAUUGAUAGACUCAAUUAUGGCAAU ..((((.--------.((((..-.((((((((....--.)))).((.(((...))))).)-)---))..)))).(((..(((((........)))))....))))))). ( -27.50) >DroPse_CAF1 49361 87 - 1 AUU----------------CGUAUUCAUGUGCAAUA--ACGGAAAGUGUCAAAGACCCAA-C---CAUAGCUCACUGUGGGGUCAAUUGAUAGACUCAAUUAUGACGAU .((----------------(((((((....).))).--)))))...(((((..(((((..-.---(((((....))))))))))((((((.....)))))).))))).. ( -21.50) >DroWil_CAF1 64454 90 - 1 AUU----------------UGUAUUCGUGUACAAUAAAACGGAAAGUGUCAAAGACCCAGUC---CAUAGCUCACCGUGGGGUCAAUUGAUAGACUCAAUUAUGAUGAU ..(----------------(((((....))))))......(((..(.(((...))).)..))---).....(((.(((((((((........)))))...)))).))). ( -16.50) >DroAna_CAF1 46200 104 - 1 AUUCGUAUUCGUGAUC--UCGUAUUCAUGUGCAAUA--ACAGAGAGUGUCAAAGACCCAA-CCACCAUAGCUCCCAGCGGGGUCAAUUGAUAGACUCAAUUAUGACGAU ..(((...(((((((.--..((((....))))....--...(((..((((((.(((((..-........((.....)).)))))..))))))..))).)))))))))). ( -24.30) >DroPer_CAF1 51212 87 - 1 AUU----------------CGUAUUCAUGUGCAAUA--ACGGAAAGUGUCAAAGACCCAA-C---CAUAGCUCACUGUGGGGUCAAUUGAUAGACUCAAUUAUGACGAU .((----------------(((((((....).))).--)))))...(((((..(((((..-.---(((((....))))))))))((((((.....)))))).))))).. ( -21.50) >consensus AUU________________CGUAUUCAUGUGCAAUA__ACGGAAAGUGUCAAAGACCCAA_C___CAUAGCUCACUGCGGGGUCAAUUGAUAGACUCAAUUAUGACGAU ....................((((....))))..............(((((..(((((...........((.....)).)))))((((((.....)))))).))))).. (-15.47 = -15.17 + -0.30)

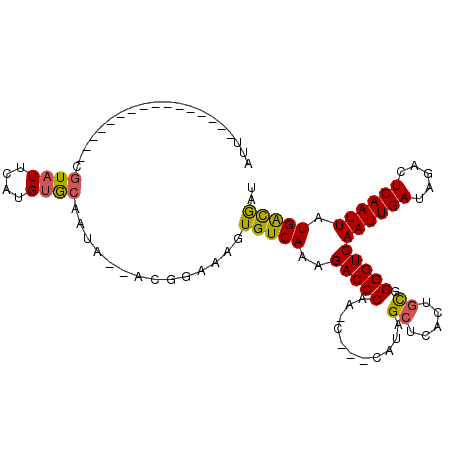

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:02 2006