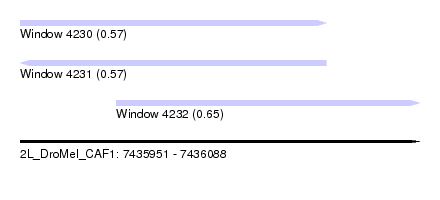
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,435,951 – 7,436,088 |
| Length | 137 |
| Max. P | 0.645736 |

| Location | 7,435,951 – 7,436,056 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -19.73 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7435951 105 + 22407834 UA-----CCCGAACUCAUAUUCUGAACCGGACGAAGUUUACC-AAUCAGGCA----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUGACGUCGGAAG ..-----............((((((...((((((((((....-.....(((.----...)))((((((......))))))))))))).)))..((((..---..))))...)))))). ( -28.70) >DroSec_CAF1 31845 105 + 1 UA-----CCCGAACUCAUAUUCUGAACCGGACGAAGUUUACC-AAUCAGGCA----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUAAAGUCGGAAG ..-----............((((((...((((((((((....-.....(((.----...)))((((((......))))))))))))).)))...(((..---..)))....)))))). ( -25.70) >DroSim_CAF1 33475 97 + 1 -------------CUCAUAUUCUGAACCGGACGAAGUUUACC-AAUCAGGCA----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUGACGUCGGAAG -------------......((((((...((((((((((....-.....(((.----...)))((((((......))))))))))))).)))..((((..---..))))...)))))). ( -28.70) >DroEre_CAF1 34359 114 + 1 CAUGUCUCCCGAACUCAUAUUCUUUACUGGACGAAGUUUACC-AAUCAGGCAUACAUAUGUCGAGCUGAUACCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUGACGUCGGAAG ........((((..(((.....(((((((((.((.((...((-.....))...))....(((((((((......))))))))).....)).))))))))---)...)))..))))... ( -34.90) >DroYak_CAF1 32325 108 + 1 UA-----CCAGAACUCAUGCACUGAACCGGACGAAGUUUACC-AAUCAGGCG----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAAUACAAACUGACGUCGGAAG ..-----.(((..(....)..)))..(((.(((.(((((.((-.....)).(----((((((((((((......))))))))).(((......)))..)))))))))..))))))... ( -27.20) >DroAna_CAF1 32415 106 + 1 UA-----CAGAAAUCCAUUCUCUCCACCAGACAAAGUUUACUUUAUUUCGUA----UAUAUCUCGCUGAUUUCCCCGCUCGACUUUGAGCCUUUAGUAA---UCGCUGACGUCGGAAG ..-----...................((.(((((((....))))........----......(((.(((((.....(((((....))))).......))---))).))).)))))... ( -14.50) >consensus UA_____CCCGAACUCAUAUUCUGAACCGGACGAAGUUUACC_AAUCAGGCA____UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA___AAGCUGACGUCGGAAG ..........................((((((((((((..........((((......))))((((((......))))))))))))).)))..((((.......)))).....))... (-19.73 = -19.82 + 0.09)


| Location | 7,435,951 – 7,436,056 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -20.97 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7435951 105 - 22407834 CUUCCGACGUCAGCUU---UUACUGGAGGAUCAAAGUCGAACUGGGGAAUCAGCUCGACAUA----UGCCUGAUU-GGUAAACUUCGUCCGGUUCAGAAUAUGAGUUCGGG-----UA ..(((((..(((.((.---..((((((.((.....(((((.((((....)))).)))))...----((((.....-))))....)).))))))..))....)))..)))))-----.. ( -34.80) >DroSec_CAF1 31845 105 - 1 CUUCCGACUUUAGCUU---UUACUGGAGGAUCAAAGUCGAACUGGGGAAUCAGCUCGACAUA----UGCCUGAUU-GGUAAACUUCGUCCGGUUCAGAAUAUGAGUUCGGG-----UA ..((((((((...((.---..((((((.((.....(((((.((((....)))).)))))...----((((.....-))))....)).))))))..)).....))).)))))-----.. ( -34.10) >DroSim_CAF1 33475 97 - 1 CUUCCGACGUCAGCUU---UUACUGGAGGAUCAAAGUCGAACUGGGGAAUCAGCUCGACAUA----UGCCUGAUU-GGUAAACUUCGUCCGGUUCAGAAUAUGAG------------- .........(((.((.---..((((((.((.....(((((.((((....)))).)))))...----((((.....-))))....)).))))))..))....))).------------- ( -27.80) >DroEre_CAF1 34359 114 - 1 CUUCCGACGUCAGCUU---UUACUGGAGGAUCAAAGUCGAACUGGGGUAUCAGCUCGACAUAUGUAUGCCUGAUU-GGUAAACUUCGUCCAGUAAAGAAUAUGAGUUCGGGAGACAUG (((((((..(((..((---((((((((.((.....(((((.((((....)))).)))))....((.((((.....-)))).)).)).))))))))))....)))..)))))))..... ( -43.90) >DroYak_CAF1 32325 108 - 1 CUUCCGACGUCAGUUUGUAUUACUGGAGGAUCAAAGUCGAACUGGGGAAUCAGCUCGACAUA----CGCCUGAUU-GGUAAACUUCGUCCGGUUCAGUGCAUGAGUUCUGG-----UA ...((((..(((...((((((((((((.((.....(((((.((((....)))).)))))...----.(((.....-))).....)).))))))..)))))))))..)).))-----.. ( -31.10) >DroAna_CAF1 32415 106 - 1 CUUCCGACGUCAGCGA---UUACUAAAGGCUCAAAGUCGAGCGGGGAAAUCAGCGAGAUAUA----UACGAAAUAAAGUAAACUUUGUCUGGUGGAGAGAAUGGAUUUCUG-----UA .((((...(((.((((---((.((....((((......))))..)).)))).))..)))...----.((.(.((((((....)))))).).))))))((((.....)))).-----.. ( -21.90) >consensus CUUCCGACGUCAGCUU___UUACUGGAGGAUCAAAGUCGAACUGGGGAAUCAGCUCGACAUA____UGCCUGAUU_GGUAAACUUCGUCCGGUUCAGAAUAUGAGUUCGGG_____UA .........(((.........((((((.((.....(((((.((((....)))).))))).......((((......))))....)).))))))........))).............. (-20.97 = -21.03 + 0.06)

| Location | 7,435,984 – 7,436,088 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7435984 104 + 22407834 UACC-AAUCAGGCA----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUGACGUCGGAAGUGCUACAACGACA-------AUGUGCAACUAAAGCCAAC ....-.....(((.----...(((((((((......)))))))))((((((....((((..---..))))..))))))(((((.(((......-------.))))).)))...)))... ( -28.30) >DroSec_CAF1 31878 104 + 1 UACC-AAUCAGGCA----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUAAAGUCGGAAGUGCUACAACGACA-------AUGUGCAACUAAAGCCAAC ....-.....(((.----...(((((((((......)))))))))................---..((....((((............)))).-------....)).......)))... ( -25.70) >DroSim_CAF1 33500 104 + 1 UACC-AAUCAGGCA----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUGACGUCGGAAGUGCUACAACGACA-------AUGUGCAACUAAAGCCAAC ....-.....(((.----...(((((((((......)))))))))((((((....((((..---..))))..))))))(((((.(((......-------.))))).)))...)))... ( -28.30) >DroEre_CAF1 34397 108 + 1 UACC-AAUCAGGCAUACAUAUGUCGAGCUGAUACCCCAGUUCGACUUUGAUCCUCCAGUAA---AAGCUGACGUCGGAAGAGCUAUAGCGGCA-------AAGUGCAGCUAAAGCCAAC ....-.....(((...((...(((((((((......)))))))))..))............---.(((((((.((....))(((.....))).-------..)).)))))...)))... ( -33.70) >DroYak_CAF1 32358 107 + 1 UACC-AAUCAGGCG----UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAAUACAAACUGACGUCGGAAGUGCCAUAACGACA-------AAGUGCAGCCAAAACCAAC ....-.....((((----((((((((((((......)))))))))..........((((.......))))..((((............)))).-------..)))).)))......... ( -28.70) >DroAna_CAF1 32448 112 + 1 UACUUUAUUUCGUA----UAUAUCUCGCUGAUUUCCCCGCUCGACUUUGAGCCUUUAGUAA---UCGCUGACGUCGGAAGUGCAGAGAACUCAUGGAGCCCCAUGGAACAAAAGUCAAC .(((((..(((...----....((((((..((((((..(((((....)))))..(((((..---..)))))....)))))))).))))...(((((....))))))))..))))).... ( -27.30) >consensus UACC_AAUCAGGCA____UAUGUCGAGCUGAUUCCCCAGUUCGACUUUGAUCCUCCAGUAA___AAGCUGACGUCGGAAGUGCUACAACGACA_______AUGUGCAACUAAAGCCAAC ..........(((........(((((((((......)))))))))((((((....((((.......))))..))))))...................................)))... (-18.02 = -18.72 + 0.70)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:01 2006