| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,414,279 – 7,414,389 |
| Length | 110 |
| Max. P | 0.629587 |

| Location | 7,414,279 – 7,414,389 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
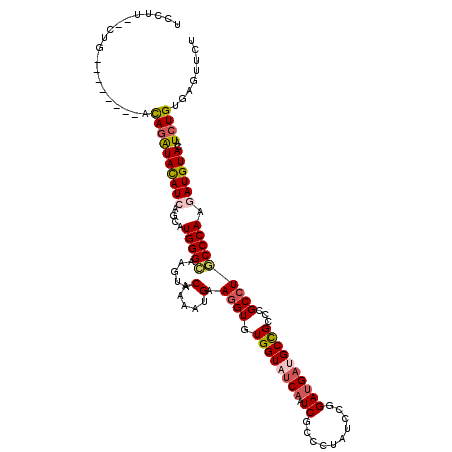
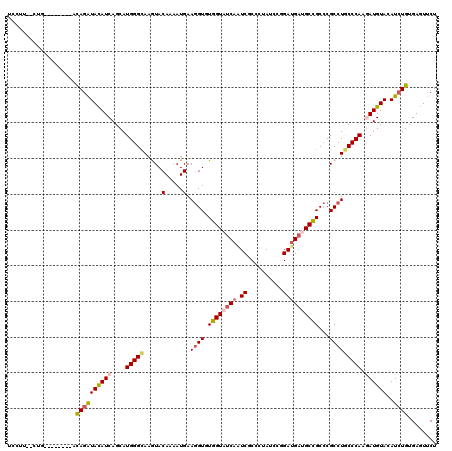
>2L_DroMel_CAF1 7414279 110 + 22407834 UACGU--CUG--------ACAGAUAUAUCAGCAUGGGCAAGUACAAGAUGAAGGUCUGGUAUCAAUCCCCUUAUCCGGACGAUGCUGCCCGCCUGCCCAAAAUGUAUAUCUGCGAAUUCU .....--.((--------.((((((((.((...((((((.((...((((....))))((((((..(((........))).))))))....)).))))))...))))))))))))...... ( -31.00) >DroVir_CAF1 6635 112 + 1 UCCAUUUUCG--------ACAGAUACAUCAGCAUGGGCAAGUACAAAAUGAAAGUGUGGUAUCAGUCGCCGUAUCCGGAUGAUGCCGCACGCCUGCCCAAGAUGUACAUCUGUGAGUUCU ......(((.--------(((((((((((....((((((.((.(.....)...(((((((((((....(((....))).))))))))))))).)))))).))))))..)))))))).... ( -42.80) >DroGri_CAF1 8696 112 + 1 UCAUUCUUAC--------ACAGAUAUAUCAGCAUGGGCAAGUACAAAAUGAAGGUGUGGUAUCAGUCGCCCUAUCCAGAUGAUGCCGCCCGCCUGCCCAAGAUGUACAUCUGUGAGUUCU .........(--------(((((((((((....((((((.((.(.....)..((.(((((((((.((..........))))))))))))))).)))))).))))))..))))))...... ( -37.60) >DroWil_CAF1 9397 107 + 1 UU-----CUC--------CCAGAUACAUCAGCAUGGGUAAAUACAAAAUGAAGGUCUGGUAUCAAUCUCCAUAUCCCGAUGAUGCUGCCCGCCUACCCAAAAUGUAUAUAUGUGAGUUCU ..-----(((--------.((.((((((.....((((((....(.....)..(((..(((((((.((..........))))))))))))....))))))..))))))...)).))).... ( -22.20) >DroMoj_CAF1 11421 107 + 1 UCCUU-------------GCAGAUACAUCAGCAUGGGCAAAUACAAAAUGAAGGUGUGGUAUCAGUCGCCCUAUCCAGAUGACGCCGCCCGCCUGCCCAAGAUGUACAUCUGUGAGUUCU ..((.-------------.((((((((((....((((((....(.....)..((.((((..(((.((..........)))))..))))))...)))))).))))))..))))..)).... ( -35.70) >DroAna_CAF1 9105 117 + 1 -UUUC--AUGCUAAUCCCUUAGGUACAUCAGCAUGGGCAAGUACAAGAUGAAGGUAUGGUACCAAUCGCCCUAUCCGGAUGACGCCGCCCGCCUCCCCAAGAUGUAUAUCUGCGAGUUCU -....--(((((...((....))......)))))((((..............((((.(((.......))).)))).((......))))))..(((....((((....))))..))).... ( -21.60) >consensus UCCUU__CUG________ACAGAUACAUCAGCAUGGGCAAGUACAAAAUGAAGGUGUGGUAUCAAUCGCCCUAUCCGGAUGAUGCCGCCCGCCUGCCCAAGAUGUACAUCUGUGAGUUCU ...................((((((((((....(((((.....(.....).((((.((((((((.((..........))))))))))...))))))))).))))))..))))........ (-20.69 = -21.25 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:51 2006