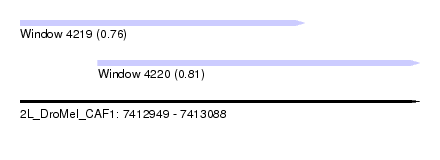
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,412,949 – 7,413,088 |
| Length | 139 |
| Max. P | 0.812833 |

| Location | 7,412,949 – 7,413,048 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -21.16 |
| Energy contribution | -22.44 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7412949 99 + 22407834 ACCGAGCAAUCCUACCCAGCGAUCGGCCAACAACAACACAAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAAUAG ..((((.........((((.((((((................)))))).)))).....(((((((((....).))))))))......))))........ ( -30.49) >DroSec_CAF1 7431 99 + 1 ACCCAGCAAUCCUACCCAGCGAUCGGCCAACAACAACACUAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAACAG ...............((((.((((((................)))))).)))).....((((((((..((((.....))))....))))))))...... ( -28.99) >DroSim_CAF1 7699 99 + 1 ACCCAGCAAUCCUACCCAGCGAUCGGCCAACAACAACACUAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAACAG ...............((((.((((((................)))))).)))).....((((((((..((((.....))))....))))))))...... ( -28.99) >DroEre_CAF1 10008 99 + 1 ACCCAGCAAUCCUACCCAGCGAUCAGCAAACAACAACACCAACCGAUCACUGGAAAACGGCGAGGGCAAACGACCCUCGCUCUCCUCCUCGUCCAACAG ...............((((.((((....................)))).)))).....(((((((((....).)))))))).................. ( -23.85) >DroYak_CAF1 8148 99 + 1 AUCCAGCAAUCCUCCCCAGCGAUCAGCAAACAACAACACCAACCGAUCACUGGAGAACGGCGAGGGCAAACGACCCUCGCUCUCCUCCUCGUCCAACAG ............((.((((.((((....................)))).)))).))..(((((((((....).)))))))).................. ( -24.95) >DroAna_CAF1 7811 96 + 1 AGCCAAUAAUCCGCCGCAACGGACGGCGGGA---AACACAAAUCGAAAUCAUGCCAAUGGCGAGGGAAAGAGGCCCUCGCUUUCAUCCUCUUCGAAUAG .........((((((((....).))))))).---........(((((...(((.....((((((((.......))))))))..)))....))))).... ( -30.80) >consensus ACCCAGCAAUCCUACCCAGCGAUCGGCCAACAACAACACAAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAACAG ...............((((.((((((................)))))).)))).....((((((((.......)))))))).................. (-21.16 = -22.44 + 1.28)


| Location | 7,412,976 – 7,413,088 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.49 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -27.13 |
| Energy contribution | -28.63 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7412976 112 + 22407834 CAACAACAACACAAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAAUAGCACAGAUGACUCUAUGAAGAUCGUUAAAAACGGGCGGAAG ........................((((...(((((((((....).))))))))...))))((((((.........(((((.(((....))))))))......))))))... ( -30.46) >DroSec_CAF1 7458 112 + 1 CAACAACAACACUAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAACAGCACGGAUGACUCUAUGAAGAUCGUUAAAAACGGCCGCAAG ............(((.(((((...((((...(((((((((....).))))))))...))))((((((........)))))).........)))))))).............. ( -34.30) >DroSim_CAF1 7726 112 + 1 CAACAACAACACUAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAACAGCACGGAUGACUCUAUGAAGAUCGUCAAAAACGGCCGCAAG .................((((...((((...(((((((((....).))))))))...))))((((((........)))))).........))))(((......)))...... ( -34.10) >DroEre_CAF1 10035 112 + 1 AAACAACAACACCAACCGAUCACUGGAAAACGGCGAGGGCAAACGACCCUCGCUCUCCUCCUCGUCCAACAGUACGGAUGACUCAAUGAAGAUCGUCAAGAACGGCCGCAAG ................(((((...(((....(((((((((....).))))))))....)))((((((........)))))).........)))))................. ( -31.10) >DroYak_CAF1 8175 112 + 1 AAACAACAACACCAACCGAUCACUGGAGAACGGCGAGGGCAAACGACCCUCGCUCUCCUCCUCGUCCAACAGUACGGAUGACUCAAUGAAGAUCGUCAAGAACGGCCGCAAG ................(((((...((((...(((((((((....).))))))))...))))((((((........)))))).........)))))................. ( -34.20) >DroAna_CAF1 7838 109 + 1 GGGA---AACACAAAUCGAAAUCAUGCCAAUGGCGAGGGAAAGAGGCCCUCGCUUUCAUCCUCUUCGAAUAGCACAGACGACUCCAUGAAGAUCGUGAAGAAUGGUCGCAAG .(..---..)...............((((..((((((((.......))))))))(((((..((((((...(((......).))...))))))..)))))...))))...... ( -27.80) >consensus CAACAACAACACAAAUCGAUCACUGGAGAACGGCGAGGGCAAGCGACCCUCGCUCUCCUCCUCGUCCAACAGCACGGAUGACUCUAUGAAGAUCGUCAAAAACGGCCGCAAG ................(((((...((((...((((((((.......))))))))...))))((((((........)))))).........)))))................. (-27.13 = -28.63 + 1.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:50 2006