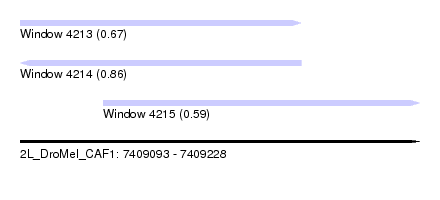
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,409,093 – 7,409,228 |
| Length | 135 |
| Max. P | 0.862809 |

| Location | 7,409,093 – 7,409,188 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -45.18 |
| Consensus MFE | -25.79 |
| Energy contribution | -27.02 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
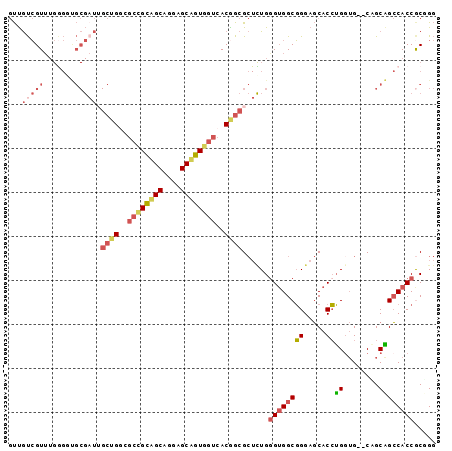
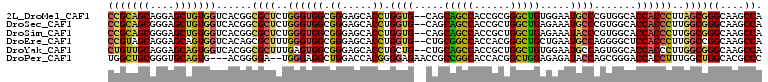
>2L_DroMel_CAF1 7409093 95 + 22407834 GUUGUCGUUUGGGGUGCGAUUGCUGGCGCCGCAGCAGGAGCUGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG--CAGCAGCCACCGCGGG ....((((...((.(((....((((..((((((((....))))))))..))))((.(..((((.(....)))))..).)--).))).))...)))). ( -48.30) >DroSec_CAF1 3605 95 + 1 GUUGUCGUUUGGGCUGCGAUUGCUGGCGCCGCAGCGGGAGCUGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG--CAGCAGCCACCGCUGG ......((...((((((....((((..((((((((....))))))))..))))((.(..((((.(....)))))..).)--).))))))...))... ( -52.10) >DroSim_CAF1 3846 95 + 1 GUUGUCGUUUGGGCUGCGAUUGCUGGCGCCGCAGCGGGAGCUGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG--CAGCAGCCACCGCUGG ......((...((((((....((((..((((((((....))))))))..))))((.(..((((.(....)))))..).)--).))))))...))... ( -52.10) >DroEre_CAF1 6000 95 + 1 GUUGUCGUUUGAGGCGCGAUGGCUGGUGCCGUAGCAGGAGCAGUGGUCACAGCGCUUUGGGUGGCGGGAGCACCUGGUG--CUGCGGCCACCACGGG ....((((..(((((((..((((((.((((.......).))).))))))..))))))).((((((.(.(((((...)))--)).).)))))))))). ( -46.20) >DroYak_CAF1 4139 95 + 1 GUUGUCGUUUGGGGUGCGCUUGCUGCGGCUGUUGCAGGAGCAGUGGUCACGGCGCUUUGAGUGGCGGGAGCACCUGCUG--CUGCAGCCACCGCUGG ..........(.(((((((.....)))...(((((((.(((((((.((.((.(((.....))).)).)).)).))))).--))))))))))).)... ( -42.60) >DroPer_CAF1 5813 77 + 1 ACCACAGU---------------GGGCGUGGCUGCGGGUGCAGUG---ACGGGGA--UGGGAGGCUGGACCACGGGGAGAACCGCCGGCACCACGGC ......((---------------((.(((.(((((....))))).---)))....--......(((((....(((......)))))))).))))... ( -29.80) >consensus GUUGUCGUUUGGGGUGCGAUUGCUGGCGCCGCAGCAGGAGCAGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG__CAGCAGCCACCGCGGG ...(((((.......))))).((((..((((((((....))))))))..))))......((((((((.....))((........))))))))..... (-25.79 = -27.02 + 1.23)


| Location | 7,409,093 – 7,409,188 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -20.96 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
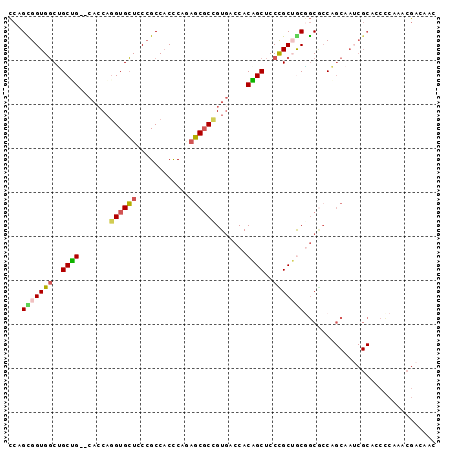
>2L_DroMel_CAF1 7409093 95 - 22407834 CCCGCGGUGGCUGCUG--CACCAGGUGCUCCCGCCACCCAGAGCGCCGUGACCACAGCUCCUGCUGCGGCGCCAGCAAUCGCACCCCAAACGACAAC ...(((((.((((...--.....(((((((..........)))))))(((.((.((((....)))).))))))))).)))))............... ( -37.10) >DroSec_CAF1 3605 95 - 1 CCAGCGGUGGCUGCUG--CACCAGGUGCUCCCGCCACCCAGAGCGCCGUGACCACAGCUCCCGCUGCGGCGCCAGCAAUCGCAGCCCAAACGACAAC ...(((..((((((((--(....(((((((..........)))))))(((.((.((((....)))).)))))..)))...))))))....)).)... ( -38.60) >DroSim_CAF1 3846 95 - 1 CCAGCGGUGGCUGCUG--CACCAGGUGCUCCCGCCACCCAGAGCGCCGUGACCACAGCUCCCGCUGCGGCGCCAGCAAUCGCAGCCCAAACGACAAC ...(((..((((((((--(....(((((((..........)))))))(((.((.((((....)))).)))))..)))...))))))....)).)... ( -38.60) >DroEre_CAF1 6000 95 - 1 CCCGUGGUGGCCGCAG--CACCAGGUGCUCCCGCCACCCAAAGCGCUGUGACCACUGCUCCUGCUACGGCACCAGCCAUCGCGCCUCAAACGACAAC ..(((((((((.(.((--(((...))))).).))))))....((((.(((......(((..(((....)))..)))))).)))).....)))..... ( -35.00) >DroYak_CAF1 4139 95 - 1 CCAGCGGUGGCUGCAG--CAGCAGGUGCUCCCGCCACUCAAAGCGCCGUGACCACUGCUCCUGCAACAGCCGCAGCAAGCGCACCCCAAACGACAAC ...(((((.(.(((((--.((((((((((............))))))((....)))))).))))).).))))).((....))............... ( -31.90) >DroPer_CAF1 5813 77 - 1 GCCGUGGUGCCGGCGGUUCUCCCCGUGGUCCAGCCUCCCA--UCCCCGU---CACUGCACCCGCAGCCACGCCC---------------ACUGUGGU ((((..(((..(((((((..((....))...)))).....--.....((---..((((....))))..))))))---------------))..)))) ( -25.30) >consensus CCAGCGGUGGCUGCUG__CACCAGGUGCUCCCGCCACCCAGAGCGCCGUGACCACAGCUCCCGCUGCGGCGCCAGCAAUCGCACCCCAAACGACAAC ...(((((((..((((.......((((((............)))))).......))))..))))))).............................. (-20.96 = -20.66 + -0.30)

| Location | 7,409,121 – 7,409,228 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -52.75 |
| Consensus MFE | -36.99 |
| Energy contribution | -36.33 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
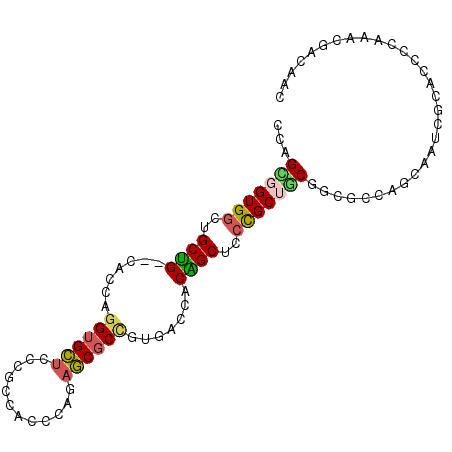
>2L_DroMel_CAF1 7409121 107 + 22407834 CCGCAGCAGGAGCUGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG--CAGCAGCCACCGCGGGCUGUGGAAUGCCCGUGGCACCACCCUUAGCGGGCAAGCCA ..(((.((((.(((((....)))))(((((.(....).))))).)))).))--).((((((......))))))((..(((((((((......))...)))))))..)). ( -54.50) >DroSec_CAF1 3633 107 + 1 CCGCAGCGGGAGCUGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG--CAGCAGCCACCGCUGGCUGAGAAAUGCCCGUGGCACCACCCUUGGCGGGCAAGCCA (((((((....)))))))....(((((.(..((((.(....)))))..).)--)..((((((....)))))).....(((((((((......))...))))))).))). ( -55.70) >DroSim_CAF1 3874 107 + 1 CCGCAGCGGGAGCUGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG--CAGCAGCCACCGCUGGCUGAGAAAUACCCGUGGCACCACCCUUGGCGGGCAAGCCA (((((((....)))))))......((((..((((((((((.((((...)))--)..((((((....)))))).......)))).....))))))..))))((....)). ( -52.80) >DroEre_CAF1 6028 107 + 1 CCGUAGCAGGAGCAGUGGUCACAGCGCUUUGGGUGGCGGGAGCACCUGGUG--CUGCGGCCACCACGGGCUGCUGAAUGCCAGGGGCUCCACCCUUGGCCGGCAAGCCA .(((.((....)).((((((.(((((((..(((((.(....))))))))))--))).)))))).)))((((((((...((((((((.....)))))))))))).)))). ( -59.60) >DroYak_CAF1 4167 107 + 1 CUGUUGCAGGAGCAGUGGUCACGGCGCUUUGAGUGGCGGGAGCACCUGCUG--CUGCAGCCACCGCUGGCUGUGGAAUGCCAGUGGCACCACCCUUGGCGGGCAAGCCA ..(((((((.(((((((.((.((.(((.....))).)).)).)).))))).--)))))))..((((((((........)))))))).........((((......)))) ( -53.00) >DroPer_CAF1 5826 104 + 1 UGGCUGCGGGUGCAGUG---ACGGGGA--UGGGAGGCUGGACCACGGGGAGAACCGCCGGCACCACGGCUGGAGAGAUACCAGCGGGACCACCUUUGGCUGGCACGCCC .((((((.(((.(((..---..((((.--(((...(((((....(((......)))))))).)))(.(((((.......))))).)..)).)).)))))).))).))). ( -40.90) >consensus CCGCAGCAGGAGCAGUGGUCACGGCGCUCUGGGUGGCGGGAGCACCUGGUG__CAGCAGCCACCGCGGGCUGAGAAAUGCCAGUGGCACCACCCUUGGCGGGCAAGCCA (((((((....)))))))......((((..((((((.((.....)).((((.....(((((......))))).....)))).......))))))..))))((....)). (-36.99 = -36.33 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:45 2006