| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,401,931 – 7,402,028 |
| Length | 97 |
| Max. P | 0.953518 |

| Location | 7,401,931 – 7,402,028 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
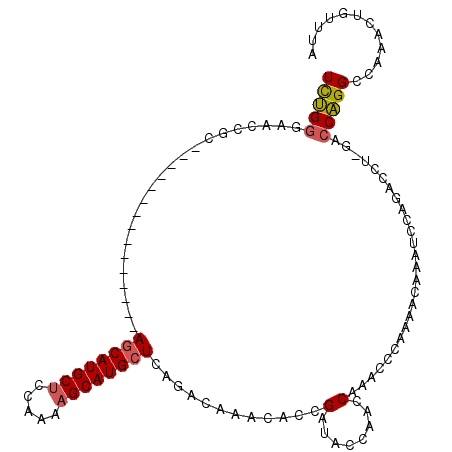
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7401931 97 + 22407834 UAAACAGUUUGGCCUGGUC-AGGUCUGGAUUUGUUUUGGGUUUGGUCGGUAUCGGUGUGUGUCUGAGCAUGCUUUUGGAGCAUGCU---------------GCGGUUCCCAGA .......(((((.(((..(-((..(..((.....))..)..)))..))).............((((((((((((...)))))))))---------------.)))...))))) ( -30.90) >DroSec_CAF1 13836 97 + 1 UAAACAGUUUGGCCUGGUC-AGGUCUGGAUUUGUUUUGGGUUUGGUUGGUAUCGGUGUUUGUCUGAGCAUGCUUUUGGAGCAUGCU---------------GCGGUUCCCAGA .((((((((..((((....-))).)..)).))))))((((..(.((.....((((.......))))((((((((...)))))))).---------------)).)..)))).. ( -30.20) >DroSim_CAF1 13942 97 + 1 UAAACAGUUUGGCCUGGUC-AGGUCUGGAUUUGUUUUGGGUUUGGUUGGUAUCGGUGUUUGUCUGAGCAUGCUUUUGGAGCAUGCU---------------GCGGUUCCCAGA .((((((((..((((....-))).)..)).))))))((((..(.((.....((((.......))))((((((((...)))))))).---------------)).)..)))).. ( -30.20) >DroEre_CAF1 14339 97 + 1 UAAACAGUUUGGCCUGGUC-AGGUCUCUAUUUGUUUUGGGUUUGGACGGUAUCGGUGUUUGUCUGAGCAUGCUUUUGGAGCAUGCU---------------GCGCUUCCCAGA .((((((...(((((....-))))).....))))))((((.(..(((((.........)))))..)((((((((...)))))))).---------------......)))).. ( -30.10) >DroAna_CAF1 16485 112 + 1 UAAACAGUUUGGCCUGGUC-AGGUCUGGAUUUGUUUUGGGUCUGGCUAGUGGCUUUGUACAUUGGAGCAUGCUUUCGAAGCAUGCUUUCAAAGCAUGCCUUGGGGUAACCAGA ..........(((((((((-((..(..((.....))..)..))))))))..((((((......((((((((((.....))))))))))))))))..)))((((.....)))). ( -45.00) >DroPer_CAF1 14608 83 + 1 UAAACAAUUUGGCCAGGUCUAGGCCU-----------CGGUCUGGCU---GCCUGUCUGCGUCUGAGCAUGCAUUUGGGGCAUGUU----------------CUAUACACCAA ........((((.(((((((((((..-----------..))))))..---))))).....((..((((((((.......)))))))----------------)...)).)))) ( -28.00) >consensus UAAACAGUUUGGCCUGGUC_AGGUCUGGAUUUGUUUUGGGUUUGGUUGGUAUCGGUGUUUGUCUGAGCAUGCUUUUGGAGCAUGCU_______________GCGGUUCCCAGA ..........(((((.....))))).........((((((...(((....)))............(((((((((...))))))))).....................)))))) (-17.99 = -18.80 + 0.81)

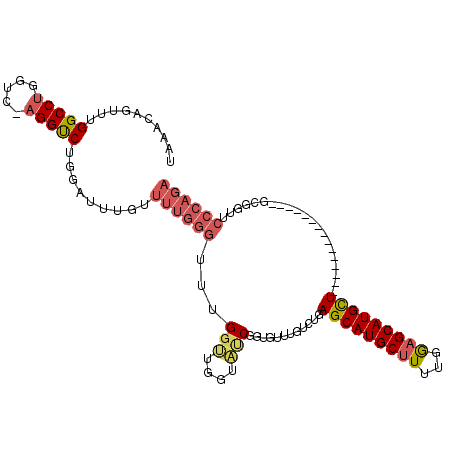
| Location | 7,401,931 – 7,402,028 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -11.73 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7401931 97 - 22407834 UCUGGGAACCGC---------------AGCAUGCUCCAAAAGCAUGCUCAGACACACACCGAUACCGACCAAACCCAAAACAAAUCCAGACCU-GACCAGGCCAAACUGUUUA ((((((....(.---------------((((((((.....)))))))))..........((....)).................))))))...-...(((......))).... ( -20.70) >DroSec_CAF1 13836 97 - 1 UCUGGGAACCGC---------------AGCAUGCUCCAAAAGCAUGCUCAGACAAACACCGAUACCAACCAAACCCAAAACAAAUCCAGACCU-GACCAGGCCAAACUGUUUA ((((((....(.---------------((((((((.....)))))))))...........(....)..................))))))...-...(((......))).... ( -18.90) >DroSim_CAF1 13942 97 - 1 UCUGGGAACCGC---------------AGCAUGCUCCAAAAGCAUGCUCAGACAAACACCGAUACCAACCAAACCCAAAACAAAUCCAGACCU-GACCAGGCCAAACUGUUUA ((((((....(.---------------((((((((.....)))))))))...........(....)..................))))))...-...(((......))).... ( -18.90) >DroEre_CAF1 14339 97 - 1 UCUGGGAAGCGC---------------AGCAUGCUCCAAAAGCAUGCUCAGACAAACACCGAUACCGUCCAAACCCAAAACAAAUAGAGACCU-GACCAGGCCAAACUGUUUA ..((((....(.---------------((((((((.....))))))))).(((.............)))....))))....((((((.(.(((-....))).)...)))))). ( -22.62) >DroAna_CAF1 16485 112 - 1 UCUGGUUACCCCAAGGCAUGCUUUGAAAGCAUGCUUCGAAAGCAUGCUCCAAUGUACAAAGCCACUAGCCAGACCCAAAACAAAUCCAGACCU-GACCAGGCCAAACUGUUUA ((((((((......(((.(((.(((..((((((((.....)))))))).))).)))....)))..))))))))....(((((......(.(((-....)))).....))))). ( -28.70) >DroPer_CAF1 14608 83 - 1 UUGGUGUAUAG----------------AACAUGCCCCAAAUGCAUGCUCAGACGCAGACAGGC---AGCCAGACCG-----------AGGCCUAGACCUGGCCAAAUUGUUUA ...((((...(----------------(.(((((.......))))).))..))))((((((..---.(((((..(.-----------.......)..)))))....)))))). ( -22.40) >consensus UCUGGGAACCGC_______________AGCAUGCUCCAAAAGCAUGCUCAGACAAACACCGAUACCAACCAAACCCAAAACAAAUCCAGACCU_GACCAGGCCAAACUGUUUA (((((......................((((((((.....))))))))............(........)..........................)))))............ (-11.73 = -11.82 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:41 2006