
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,393,059 – 7,393,153 |
| Length | 94 |
| Max. P | 0.970506 |

| Location | 7,393,059 – 7,393,153 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
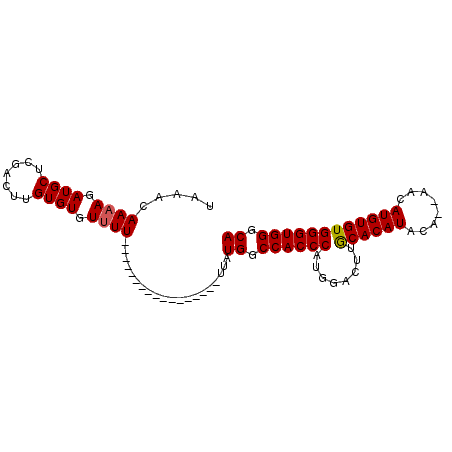
>2L_DroMel_CAF1 7393059 94 + 22407834 UAAACAAAAGAUGCUCGACUUGUGUGUGUUUUUUUUUUGUUUUUUUGUUAUGGCCACCCAUUGACUUGCACAUACA--UACAUGUGUGGGUGGGCA .((((((((((.((.(.((....))).))...))))))))))........((.((((((........((((((...--...)))))))))))).)) ( -25.40) >DroSec_CAF1 4603 77 + 1 UAAACAAAAGAUGCUCGACUUGUGUGUUUU-----------------UUAUGGCCACCCAUGGACUUGCACAUACA--AACAUGUGUGGGUGGGCA .....(((((((((.(.....).)))))))-----------------)).((.((((((........((((((...--...)))))))))))).)) ( -22.40) >DroSim_CAF1 4603 77 + 1 UAAACAAAAGAUGCUCGACUUGUGUGUUUU-----------------UUAUGGCCACCCAUGGACUUGCACAUACA--AACAUGUGUGGGUGGGCA .....(((((((((.(.....).)))))))-----------------)).((.((((((........((((((...--...)))))))))))).)) ( -22.40) >DroEre_CAF1 4700 79 + 1 UAAACAAAAGAUGCUCGACUUGUGUGUUUU-----------------UUAUGGCCACCCAUGGACUAACACAUACUCCUACAUGUGUGGGUGGGCA ...........((((((...((((((((.(-----------------((((((....)))))))..))))))))..(((((....))))))))))) ( -25.40) >consensus UAAACAAAAGAUGCUCGACUUGUGUGUUUU_________________UUAUGGCCACCCAUGGACUUGCACAUACA__AACAUGUGUGGGUGGGCA .....((((.((((.......)))).))))....................((.((((((........((((((........)))))))))))).)) (-19.74 = -19.80 + 0.06)

| Location | 7,393,059 – 7,393,153 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -19.16 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7393059 94 - 22407834 UGCCCACCCACACAUGUA--UGUAUGUGCAAGUCAAUGGGUGGCCAUAACAAAAAAACAAAAAAAAAACACACACAAGUCGAGCAUCUUUUGUUUA ((.(((((((.((.((((--(....))))).))...))))))).))........((((((((.......((......))........)))))))). ( -19.96) >DroSec_CAF1 4603 77 - 1 UGCCCACCCACACAUGUU--UGUAUGUGCAAGUCCAUGGGUGGCCAUAA-----------------AAAACACACAAGUCGAGCAUCUUUUGUUUA ((.(((((((((((((..--..))))))........))))))).))...-----------------..............(((((.....))))). ( -18.70) >DroSim_CAF1 4603 77 - 1 UGCCCACCCACACAUGUU--UGUAUGUGCAAGUCCAUGGGUGGCCAUAA-----------------AAAACACACAAGUCGAGCAUCUUUUGUUUA ((.(((((((((((((..--..))))))........))))))).))...-----------------..............(((((.....))))). ( -18.70) >DroEre_CAF1 4700 79 - 1 UGCCCACCCACACAUGUAGGAGUAUGUGUUAGUCCAUGGGUGGCCAUAA-----------------AAAACACACAAGUCGAGCAUCUUUUGUUUA ((.((((((((....)).(((.((.....)).)))..)))))).))...-----------------..............(((((.....))))). ( -19.30) >consensus UGCCCACCCACACAUGUA__UGUAUGUGCAAGUCCAUGGGUGGCCAUAA_________________AAAACACACAAGUCGAGCAUCUUUUGUUUA ((.(((((((((((((......))))))........))))))).))..................................(((((.....))))). (-18.90 = -18.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:38 2006