
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,374,308 – 7,374,442 |
| Length | 134 |
| Max. P | 0.920817 |

| Location | 7,374,308 – 7,374,402 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.14 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7374308 94 + 22407834 CACGGCCAUGUCGACCGUGAUUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUG------UCAGCGCUGCAUAAGGCAUUUCAAAUAAUGCACUUGAA (((((.(.....).)))))......(.((((....((((((((.........))))------)))).))))).(((((((((......))))).)))).. ( -28.30) >DroPse_CAF1 194227 94 + 1 CAUGGCCAUGUCGGCCAUGAUUAACGUCAAC-UUUUUGACAGCCAUUUGACAGCUGCUGUUGCUAUUGCUGCAUAAGGCAUUUAAAAUAUCAUAU----- (((((((.....)))))))......(((((.-...))))).(((...((.((((.((....))....))))))...)))................----- ( -30.30) >DroEre_CAF1 163514 94 + 1 CACGGCCAUGUCGACCGUGAUUAACGUCAGCUUUUUUGACAGCCAUUUGACAGCUG------UCAGCGCUGCAUAAGGCAUUUCAAAAAAUGCACUUGAA (((((.(.....).)))))......(.((((....((((((((.........))))------)))).))))).((((((((((....)))))).)))).. ( -28.40) >DroYak_CAF1 167108 94 + 1 CACGGCCAUGUCGACCGUGAUUAACGUCAGCUUUUUUGACAGCCAUUUGACAGCUG------UCAGCGCUGCAUAAGGCAUUUCAAAAAAUGCACUUGAA (((((.(.....).)))))......(.((((....((((((((.........))))------)))).))))).((((((((((....)))))).)))).. ( -28.40) >DroAna_CAF1 171294 93 + 1 CACGGCCAUGUCGGCCAUGAUUAACGUCAACAUUUUUGACAGCCAUUUGACAGCUG------CCACCGCUGCAUAAGGCAUUUCCACAAAUACACUUUG- ((.((((.....)))).))......(((((.....))))).(((...((.((((..------.....))))))...)))....................- ( -23.60) >DroPer_CAF1 196387 94 + 1 CAUGGCCAUGUCGGCCAUGAUUAACGUCAAC-UUUUUGACAGCCAUUUGACAGCUGCUGUUGCUAUUGCUGCAUAAGGCAUUUCAAAUAUCAUAU----- (((((((.....)))))))......(((((.-...))))).(((...((.((((.((....))....))))))...)))................----- ( -30.30) >consensus CACGGCCAUGUCGACCAUGAUUAACGUCAAC_UUUUUGACAGCCAUUUGACAGCUG______CCAGCGCUGCAUAAGGCAUUUCAAAAAAUACACUUGA_ (((((.(.....).)))))......(((((.....))))).(((...((.((((.............))))))...)))..................... (-20.55 = -20.14 + -0.42)


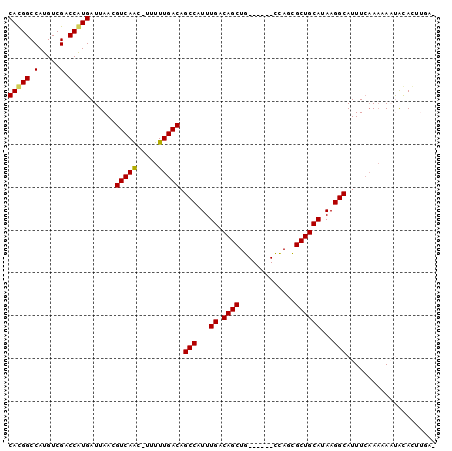
| Location | 7,374,308 – 7,374,402 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -24.19 |
| Energy contribution | -23.47 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7374308 94 - 22407834 UUCAAGUGCAUUAUUUGAAAUGCCUUAUGCAGCGCUGA------CAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAAUCACGGUCGACAUGGCCGUG ...(((.(((((......))))))))((((((((.(((------(((((((...))))))))))....))))).)))....(((((((.....))))))) ( -33.00) >DroPse_CAF1 194227 94 - 1 -----AUAUGAUAUUUUAAAUGCCUUAUGCAGCAAUAGCAACAGCAGCUGUCAAAUGGCUGUCAAAAA-GUUGACGUUAAUCAUGGCCGACAUGGCCAUG -----...((((.........(((...((((((....((....)).)))).))...))).(((((...-.)))))))))..(((((((.....))))))) ( -26.30) >DroEre_CAF1 163514 94 - 1 UUCAAGUGCAUUUUUUGAAAUGCCUUAUGCAGCGCUGA------CAGCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAAUCACGGUCGACAUGGCCGUG ...(((.((((((....)))))))))(((((((..(((------(((((((...)))))))))).....)))).)))....(((((((.....))))))) ( -33.70) >DroYak_CAF1 167108 94 - 1 UUCAAGUGCAUUUUUUGAAAUGCCUUAUGCAGCGCUGA------CAGCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAAUCACGGUCGACAUGGCCGUG ...(((.((((((....)))))))))(((((((..(((------(((((((...)))))))))).....)))).)))....(((((((.....))))))) ( -33.70) >DroAna_CAF1 171294 93 - 1 -CAAAGUGUAUUUGUGGAAAUGCCUUAUGCAGCGGUGG------CAGCUGUCAAAUGGCUGUCAAAAAUGUUGACGUUAAUCAUGGCCGACAUGGCCGUG -..(((.((((((....)))))))))((((((((.(((------(((((((...))))))))))....))))).)))....(((((((.....))))))) ( -32.00) >DroPer_CAF1 196387 94 - 1 -----AUAUGAUAUUUGAAAUGCCUUAUGCAGCAAUAGCAACAGCAGCUGUCAAAUGGCUGUCAAAAA-GUUGACGUUAAUCAUGGCCGACAUGGCCAUG -----........(((((...(((...((((((....((....)).)))).))...)))..)))))..-............(((((((.....))))))) ( -26.60) >consensus _UCAAGUGCAUUAUUUGAAAUGCCUUAUGCAGCGCUGA______CAGCUGUCAAAUGGCUGUCAAAAA_GCUGACGUUAAUCACGGCCGACAUGGCCGUG .....................(((...((((((.............)))).))...))).((((.......))))......(((((((.....))))))) (-24.19 = -23.47 + -0.72)


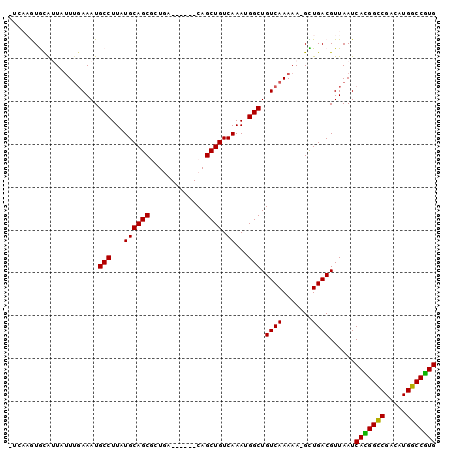
| Location | 7,374,328 – 7,374,442 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -18.81 |
| Energy contribution | -20.43 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7374328 114 + 22407834 UUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUCAGCGCUGCAUAAGGCAUUUCAAAUAAUGCACUUGAACAAUUUUCAAGAGGUUUCCCUAUCAAAUAUUUAUGAAAAU .......((((....((((((((.........)))))))).))))(((((((((((......))))).((((((.....))))))(((....)))........))))))..... ( -27.10) >DroSec_CAF1 159824 114 + 1 UUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUCAGCGCUGCAUAAGGCAUUUCAAAUAAUGCACUUGAACAAUUUUCAAGGGGGUUCUAUAACAAAUAUUUAUGAUAAU ..(((.((.((((((((((..(((...((.((((.......))))))...)))...))))).))))).((((((.....)))))))).)))....................... ( -26.60) >DroSim_CAF1 166903 113 + 1 UUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUCAGCGCUGCAUAAGGCAUUUCAAAUAAUGCACUUGAACAAUUUUCAAGGGGGUUCUUUAUCAAAUAUUUAUGAUAA- ..(((.((.((((((((((..(((...((.((((.......))))))...)))...))))).))))).((((((.....)))))))).)))..((((((........))))))- ( -28.80) >DroEre_CAF1 163534 110 + 1 UUAACGUCAGCUUUUUUGACAGCCAUUUGACAGCUGUCAGCGCUGCAUAAGGCAUUUCAAAAAAUGCACUUGAACAAUUUUCAGGGGU-UUCCUUACCAAAUAUUUAUGAU--- .......((((....((((((((.........)))))))).))))((((((((((((....)))))).((((((.....))))))(((-......))).....))))))..--- ( -28.30) >DroYak_CAF1 167128 114 + 1 UUAACGUCAGCUUUUUUGACAGCCAUUUGACAGCUGUCAGCGCUGCAUAAGGCAUUUCAAAAAAUGCACUUGAACAUUUUUCAAGGCUCUUCCUGAUCGAAUAUUUAUGAUAAU ....((((((.((((((((..(((...((.((((.......))))))...)))...)))))))).((.((((((.....)))))))).....)))).))............... ( -27.90) >DroAna_CAF1 171314 97 + 1 UUAACGUCAACAUUUUUGACAGCCAUUUGACAGCUGCCACCGCUGCAUAAGGCAUUUCCACAAAUACACUUUG------------GGGUUUG-----CAAUUUUUCGUAAUAAU .....(((((.....))))).(((...((.((((.......))))))...)))...(((.((((.....))))------------))).(((-----(........)))).... ( -19.40) >consensus UUAACGUCAGCAUUUUUGACAGCCAUUUGACAGCUGUCAGCGCUGCAUAAGGCAUUUCAAAAAAUGCACUUGAACAAUUUUCAAGGGGGUUCCUUAUCAAAUAUUUAUGAUAAU .......((((....((((((((.........)))))))).))))(((((((((((......))))).((((((.....))))))..................))))))..... (-18.81 = -20.43 + 1.63)

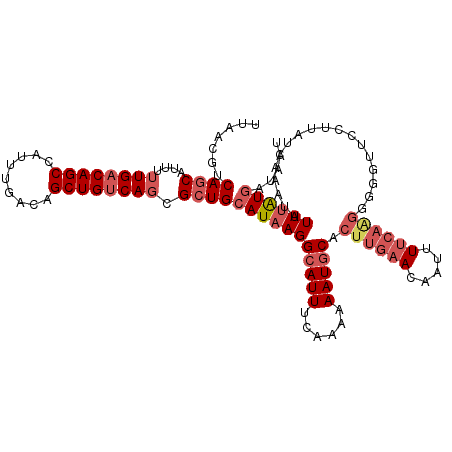
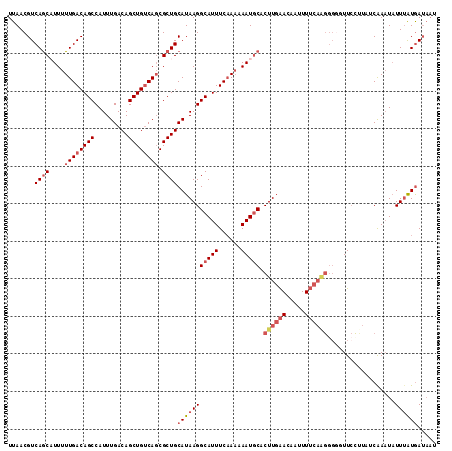
| Location | 7,374,328 – 7,374,442 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.61 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7374328 114 - 22407834 AUUUUCAUAAAUAUUUGAUAGGGAAACCUCUUGAAAAUUGUUCAAGUGCAUUAUUUGAAAUGCCUUAUGCAGCGCUGACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAA .........(((.((..(.(((....))).)..)).)))....(((.(((((......))))))))((((((((.((((((((((...))))))))))....))))).)))... ( -31.50) >DroSec_CAF1 159824 114 - 1 AUUAUCAUAAAUAUUUGUUAUAGAACCCCCUUGAAAAUUGUUCAAGUGCAUUAUUUGAAAUGCCUUAUGCAGCGCUGACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAA ................((((.........((((((.....)))))).(((((.(((((...(((...((((((.......)))).))...)))..))))))))))))))..... ( -27.00) >DroSim_CAF1 166903 113 - 1 -UUAUCAUAAAUAUUUGAUAAAGAACCCCCUUGAAAAUUGUUCAAGUGCAUUAUUUGAAAUGCCUUAUGCAGCGCUGACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAA -((((((........))))))..(((...((((((.....)))))).(((((.(((((...(((...((((((.......)))).))...)))..))))))))))....))).. ( -29.00) >DroEre_CAF1 163534 110 - 1 ---AUCAUAAAUAUUUGGUAAGGAA-ACCCCUGAAAAUUGUUCAAGUGCAUUUUUUGAAAUGCCUUAUGCAGCGCUGACAGCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAA ---........(((..(((((((..-...)))(((((.(((......))).)))))....))))..)))((((..((((((((((...)))))))))).....))))....... ( -26.80) >DroYak_CAF1 167128 114 - 1 AUUAUCAUAAAUAUUCGAUCAGGAAGAGCCUUGAAAAAUGUUCAAGUGCAUUUUUUGAAAUGCCUUAUGCAGCGCUGACAGCUGUCAAAUGGCUGUCAAAAAAGCUGACGUUAA .....(((((.((((...((((((((.((((((((.....)))))).)).))))))))))))..)))))((((..((((((((((...)))))))))).....))))....... ( -31.70) >DroAna_CAF1 171314 97 - 1 AUUAUUACGAAAAAUUG-----CAAACCC------------CAAAGUGUAUUUGUGGAAAUGCCUUAUGCAGCGGUGGCAGCUGUCAAAUGGCUGUCAAAAAUGUUGACGUUAA ......(((.......(-----((...((------------((((.....)))).))...)))......(((((.((((((((((...))))))))))....))))).)))... ( -23.30) >consensus AUUAUCAUAAAUAUUUGAUAAGGAAACCCCUUGAAAAUUGUUCAAGUGCAUUAUUUGAAAUGCCUUAUGCAGCGCUGACAGCUGUCAAAUGGCUGUCAAAAAUGCUGACGUUAA ...........................................(((.(((((......))))))))((((((((.((((((((((...))))))))))....))))).)))... (-22.38 = -22.30 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:35 2006