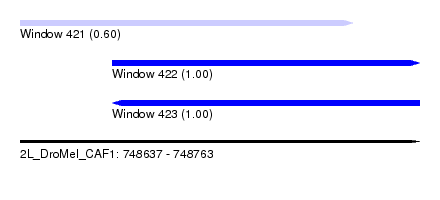
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 748,637 – 748,763 |
| Length | 126 |
| Max. P | 0.999883 |

| Location | 748,637 – 748,742 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.96 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 748637 105 + 22407834 AGAAGGCAAGCAUUACUUCCUAUUUGUUAUGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUGUAGUCCAAUCUUCAUGCCGUAUUCCAAG ((((((((((((............)))).))))))))......(((((....))))).....((((((((((((((........))))..))))))))))..... ( -25.50) >DroSec_CAF1 10163 102 + 1 AGAGGGCAAGUAUUACUUCCUAUUUGAUAUGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUG---UCCAAUCUUCAAGCCGUAUUUCAAG ((((((((..(((((.........)))))))))))))......(((((....))))).....((((((((..((((.---....))))....))))))))..... ( -23.30) >DroSim_CAF1 13597 102 + 1 AGAAGGCAAGUAUUACUUCCUAUUUGAUAUGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUG---UCCAAUCUUCAAGCCGUAUUCCAAG ((((((((..(((((.........)))))))))))))......(((((....))))).....((((((((..((((.---....))))....))))))))..... ( -23.60) >DroEre_CAF1 10494 101 + 1 GCAAGGCAAGUAUUACGUCCUACUUGAUAUGCCUUCGCUC-UUGGCAAAUAUUUGUUAUUAUAAUACGGUGCAGGUG---UCCAGUCUUCAAGCCGUAUUCCAAG ((((((((..(((((.(.....).))))))))))).)).(-((((..((((.....))))..((((((((..(((..---......)))...))))))))))))) ( -24.00) >DroYak_CAF1 12878 102 + 1 AGACGGCAAGUAUUACUUCCUAUUUGAUAUGCCUUCUCUCCUCAGCAAAUAUUUGUUAUUAUAAUACGGUGCAGGUG---UCCAGUCCUCAAGCCGUAUUCCAAG (((.((((..(((((.........))))))))).)))......(((((....))))).....((((((((..(((..---......)))...))))))))..... ( -20.00) >consensus AGAAGGCAAGUAUUACUUCCUAUUUGAUAUGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUG___UCCAAUCUUCAAGCCGUAUUCCAAG ((((((((..(((((.........)))))))))))))......(((((....))))).....((((((((..((((........))))....))))))))..... (-18.28 = -18.96 + 0.68)

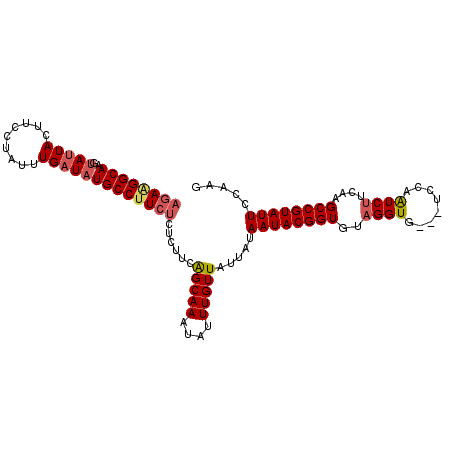
| Location | 748,666 – 748,763 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 748666 97 + 22407834 UGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUGUAGUCCAAUCUUCAUGCCGUAUUCCAAGCACCCGCCUGAGAGAAGUUAC ...((((((((...(((((....))))).....((((((((((((((........))))..))))))))))..............)))))))).... ( -24.80) >DroSec_CAF1 10192 94 + 1 UGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUG---UCCAAUCUUCAAGCCGUAUUUCAAGCACCCGCCUGAGAGAAGUUAU ...((((((((...(((((....))))).....((((((((..((((.---....))))....))))))))..............)))))))).... ( -24.90) >DroSim_CAF1 13626 94 + 1 UGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUG---UCCAAUCUUCAAGCCGUAUUCCAAGCACCCGCCUGAGAGAAGUUAC ...((((((((...(((((....))))).....((((((((..((((.---....))))....))))))))..............)))))))).... ( -24.90) >DroEre_CAF1 10523 93 + 1 UGCCUUCGCUC-UUGGCAAAUAUUUGUUAUUAUAAUACGGUGCAGGUG---UCCAGUCUUCAAGCCGUAUUCCAAGCACCCGCCUGAGAGAAGUUAC ...((((.(((-..(((.((((.....))))..((((((((..(((..---......)))...))))))))..........))).))).)))).... ( -22.60) >DroYak_CAF1 12907 94 + 1 UGCCUUCUCUCCUCAGCAAAUAUUUGUUAUUAUAAUACGGUGCAGGUG---UCCAGUCCUCAAGCCGUAUUCCAAGCACCCGCCUGAGAGAAGUUUC ...((((((((...(((((....))))).....((((((((..(((..---......)))...))))))))..............)))))))).... ( -25.00) >consensus UGCCUUCUCUCUUCAGCAAAUAUUUGUUAUUAUAAUACGGUGUAGGUG___UCCAAUCUUCAAGCCGUAUUCCAAGCACCCGCCUGAGAGAAGUUAC ...((((((((...(((((....))))).....((((((((..((((........))))....))))))))..............)))))))).... (-21.36 = -21.36 + -0.00)

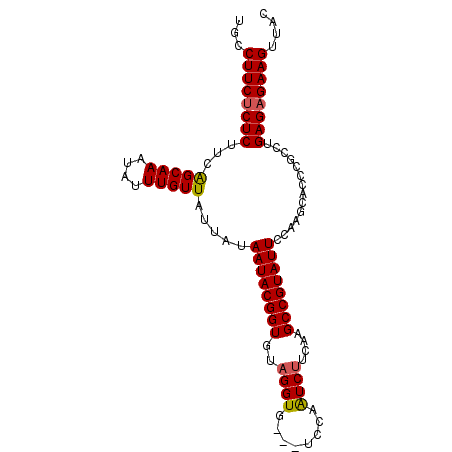
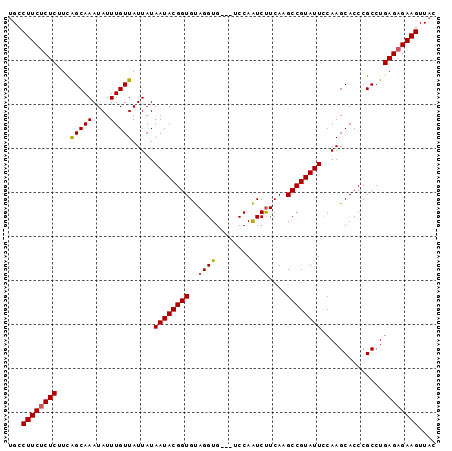
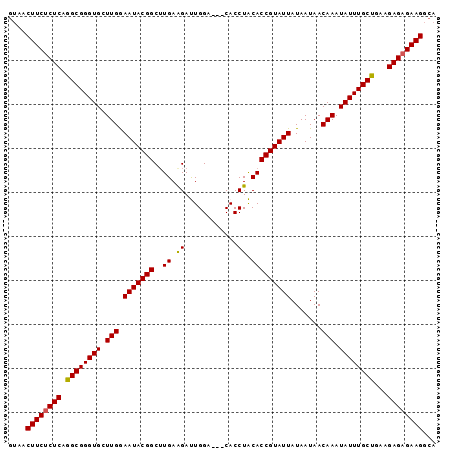
| Location | 748,666 – 748,763 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.54 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 748666 97 - 22407834 GUAACUUCUCUCAGGCGGGUGCUUGGAAUACGGCAUGAAGAUUGGACUACACCUACACCGUAUUAUAAUAACAAAUAUUUGCUGAAGAGAGAAGGCA ....((((((((.((((((((.(((.(((((((..((.((..((.....)).)).))))))))).......))).))))))))...))))))))... ( -28.60) >DroSec_CAF1 10192 94 - 1 AUAACUUCUCUCAGGCGGGUGCUUGAAAUACGGCUUGAAGAUUGGA---CACCUACACCGUAUUAUAAUAACAAAUAUUUGCUGAAGAGAGAAGGCA ....((((((((.((((((((.(((.(((((((..((.((......---...)).))))))))).......))).))))))))...))))))))... ( -25.60) >DroSim_CAF1 13626 94 - 1 GUAACUUCUCUCAGGCGGGUGCUUGGAAUACGGCUUGAAGAUUGGA---CACCUACACCGUAUUAUAAUAACAAAUAUUUGCUGAAGAGAGAAGGCA ....((((((((.((((((((.(((.(((((((..((.((......---...)).))))))))).......))).))))))))...))))))))... ( -27.80) >DroEre_CAF1 10523 93 - 1 GUAACUUCUCUCAGGCGGGUGCUUGGAAUACGGCUUGAAGACUGGA---CACCUGCACCGUAUUAUAAUAACAAAUAUUUGCCAA-GAGCGAAGGCA ....((((.(((.(((((((((..((....((((.....).)))..---..)).)))))(((((.........))))).))))..-))).))))... ( -24.80) >DroYak_CAF1 12907 94 - 1 GAAACUUCUCUCAGGCGGGUGCUUGGAAUACGGCUUGAGGACUGGA---CACCUGCACCGUAUUAUAAUAACAAAUAUUUGCUGAGGAGAGAAGGCA ....((((((((.((((((((.(((.(((((((..(((((......---..))).))))))))).......))).))))))))...))))))))... ( -32.30) >consensus GUAACUUCUCUCAGGCGGGUGCUUGGAAUACGGCUUGAAGAUUGGA___CACCUACACCGUAUUAUAAUAACAAAUAUUUGCUGAAGAGAGAAGGCA ....((((((((.((((((((.(((.(((((((..((.((............)).))))))))).......))).))))))))...))))))))... (-26.66 = -26.54 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:33 2006