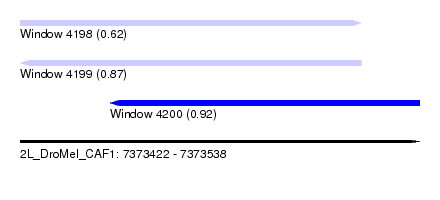
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,373,422 – 7,373,538 |
| Length | 116 |
| Max. P | 0.919294 |

| Location | 7,373,422 – 7,373,521 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -21.74 |
| Energy contribution | -23.30 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
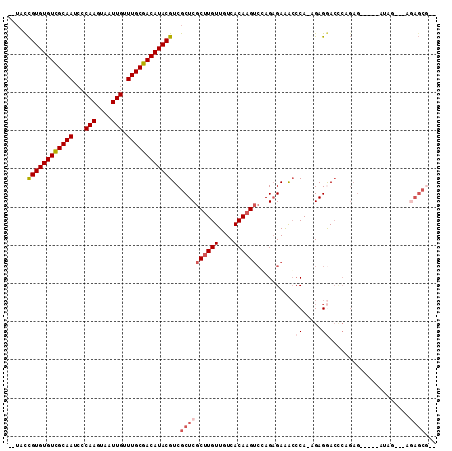
>2L_DroMel_CAF1 7373422 99 + 22407834 --CGCUCU---CUAU-----CUCUGAGUCCUCU-UGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUA-- --(((((.---....-----....(((.((...-..))...)))..(.(((((....))))))))))).(((.((((((((.(((....)))...)))))))).)))...-- ( -32.50) >DroSec_CAF1 158933 99 + 1 --CGCUCU---CUAU-----CUCUGGGUUCUCU-UGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUA-- --(((((.---....-----.((..((....))-..))........(.(((((....))))))))))).(((.((((((((.(((....)))...)))))))).)))...-- ( -31.80) >DroSim_CAF1 166002 99 + 1 --CGCUCU---CUAU-----CUCUGGGUCCUCU-UGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGUA-- --(((((.---....-----.((..((....))-..))........(.(((((....))))))))))).(((.((((((((.(((....)))...)))))))).)))...-- ( -31.80) >DroEre_CAF1 162649 99 + 1 --CGCU-----CCAG-----CUCCGGGUCCUCU-UGGGCUUCUCUGGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGGAAG --((((-----(...-----.(((((((((...-.))))....)))))(((((....))))).))))).(((.((((((((.(((....)))...)))))))).)))..... ( -36.90) >DroYak_CAF1 166212 84 + 1 ------------------------GGGUCCUCU-UGGGUUUCUCUGGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACAGG--- ------------------------..(((((((-..((....))..).(((((....))))).))).)))((.((((((((.(((....)))...)))))))).))...--- ( -24.70) >DroAna_CAF1 170309 109 + 1 AAUGCUCUUCUCCAAUAAUUCUCCUU-UUCUCUCUGUCUCUCGCUGCUCAUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCAACACACGAGA-- ..........................-.((((...(((.((((((.....(((....))))))))).)))((.(((.((((.(((....)))...)))).))).))))))-- ( -24.80) >consensus __CGCUCU___CUAU_____CUCUGGGUCCUCU_UGGGUUUCUCUCGACUUGUGACAACAAGCGAGCGACGUAUGUCGCAAACAAUUACUUGGGAUUGCGACACACGGGA__ ..(((((.................(((.(((....)))...)))....(((((....))))).))))).(((.((((((((.(((....)))...)))))))).)))..... (-21.74 = -23.30 + 1.56)

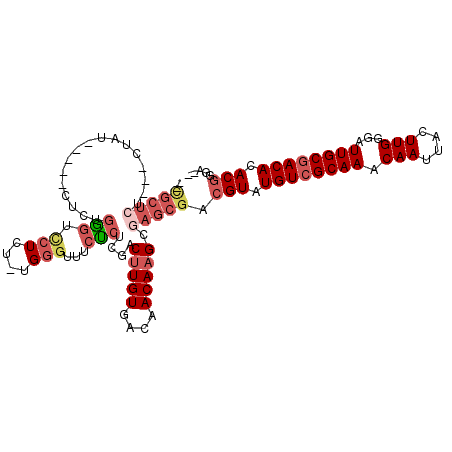
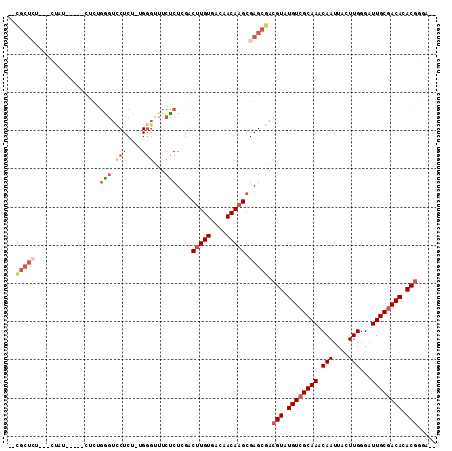
| Location | 7,373,422 – 7,373,521 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -25.16 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
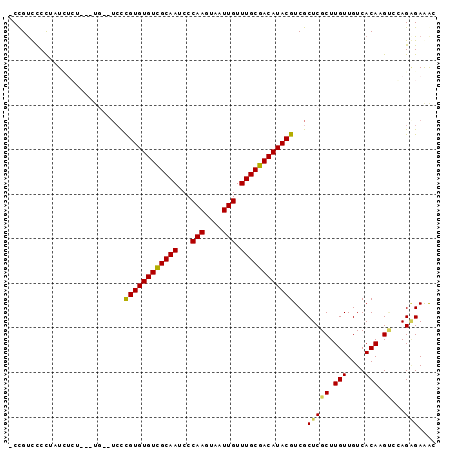
>2L_DroMel_CAF1 7373422 99 - 22407834 --UACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAACCCA-AGAGGACUCAGAG-----AUAG---AGAGCG-- --...((((((((((((...(((....))).)))))))))))).(((((((((((....))))))...(((...((..-...)).)))....-----....---.)))))-- ( -35.00) >DroSec_CAF1 158933 99 - 1 --UACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAACCCA-AGAGAACCCAGAG-----AUAG---AGAGCG-- --...((((((((((((...(((....))).)))))))))))).(((((.((((((.((.....((....))......-.)).)))...)))-----....---.)))))-- ( -32.60) >DroSim_CAF1 166002 99 - 1 --UACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAACCCA-AGAGGACCCAGAG-----AUAG---AGAGCG-- --...((((((((((((...(((....))).)))))))))))).(((((.....(((((.....((....))...((.-...)).......)-----))))---.)))))-- ( -33.40) >DroEre_CAF1 162649 99 - 1 CUUCCCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAGCCCA-AGAGGACCCGGAG-----CUGG-----AGCG-- .....((((((((((((...(((....))).)))))))))))).(((((((((((....)))))).......(((((.-.........)).)-----)).)-----))))-- ( -36.20) >DroYak_CAF1 166212 84 - 1 ---CCUGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAACCCA-AGAGGACCC------------------------ ---((((((((((((((...(((....))).)))))))))))((..(((((((((....))))))...)))..))...-..)))....------------------------ ( -29.70) >DroAna_CAF1 170309 109 - 1 --UCUCGUGUGUUGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAUGAGCAGCGAGAGACAGAGAGAA-AAGGAGAAUUAUUGGAGAAGAGCAUU --(((((((((((((((...(((....))).)))))))))))(((.((((((.(((........))))))))).))).))))...-.......................... ( -37.40) >consensus __UACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAACCCA_AGAGGACCCAGAG_____AUAG___AGAGCG__ .....((((((((((((...(((....))).))))))))))))..((((((((((....))))))..........((.....)).....................))))... (-25.16 = -26.38 + 1.22)

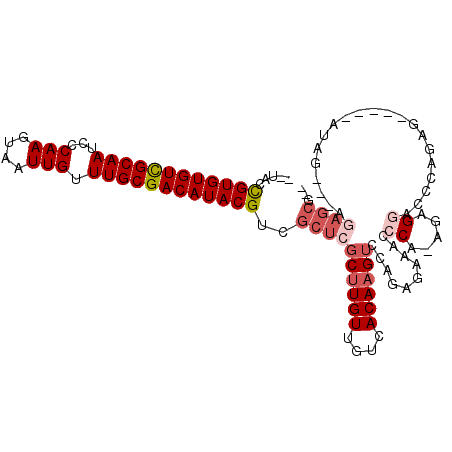
| Location | 7,373,448 – 7,373,538 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -21.11 |
| Energy contribution | -20.78 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
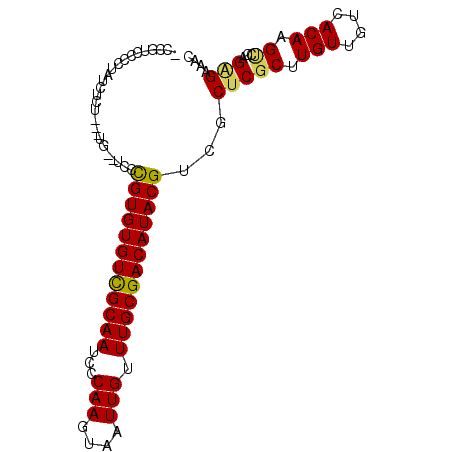
>2L_DroMel_CAF1 7373448 90 - 22407834 -CCGCUCCCCAUCGCC---UG--UACCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCGAGAGAAAC -..((........)).---..--...((((((((((((...(((....))).))))))))))))((.(((((((((....))))).)))).))... ( -32.30) >DroPse_CAF1 193297 95 - 1 -CUGUCUCCAAUGUCUCGUUGUCUCCCGUGUGUUGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAUGCAGAGAGAAAA -..........(.((((.........((((((((((((...(((....))).))))))))))))......((.(((....))).)).)))).)... ( -26.90) >DroEre_CAF1 162673 92 - 1 -CCGUUCCCUGUCUCC---UUCUUCCCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAGC -..............(---(((((..((((((((((((...(((....))).))))))))))))......((((((....))))))...)))))). ( -29.70) >DroYak_CAF1 166224 85 - 1 -CCGUUCCCUGUUC----------CCUGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAAC -...(((.(((...----------...(((((((((((...(((....))).))))))))))).......((((((....)))))).))).))).. ( -27.30) >DroAna_CAF1 170345 91 - 1 CCUGUCUCUUGUCUGU---UG--UCUCGUGUGUUGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAUGAGCAGCGAGAG .....(((((((....---..--...((((((((((((...(((....))).))))))))))))..(((((..((....))..))))).))))))) ( -32.70) >DroPer_CAF1 195446 95 - 1 -CUGUCUCCAAUGUCUCGUUGUCUCCCGUGUGUUGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAUGCAGAGGGAAAA -.......(((((...))))).....((((((((((((...(((....))).))))))))))))((.(((((.(((....))).)).))).))... ( -28.40) >consensus _CCGUCCCCUAUCUCU___UG__UCCCGUGUGUCGCAAUCCCAAGUAAUUGUUUGCGACAUACGUCGCUCGCUUGUUGUCACAAGUCCAGAGAAAC ..........................((((((((((((...(((....))).))))))))))))...(((((.(((....))).))...))).... (-21.11 = -20.78 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:31 2006