
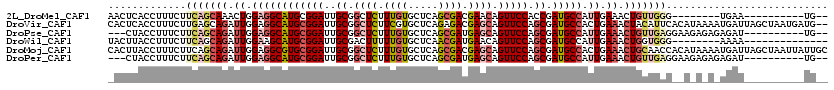
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,370,298 – 7,370,398 |
| Length | 100 |
| Max. P | 0.585325 |

| Location | 7,370,298 – 7,370,398 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -21.93 |
| Energy contribution | -23.72 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7370298 100 - 22407834 AACUCACCUUUCUUCAGCAAACUGGAGGCAUGCGGAUUGCGGCUCUUUGUGCUCAGCGACGAACAGUUCCACCGAUGCCAUUGAAACUGUUGGG--------UGAA----------UG-- ...((((((..((((((....)))))(((((.(((..((..(((.(((((((...)).))))).)))..)))))))))).........)..)))--------))).----------..-- ( -31.20) >DroVir_CAF1 211092 118 - 1 CACUCACCUUUCUUGAGCAGAUUGGAGGCAUGCGGAUUGCGGCUCUUCGUGCUCAGAGACGAGCAGUUCCAGCGAUGCCACUGAAACUACAUUCACAUAAAAUGAUUAGCUAAUGAUG-- ..........((...(((...(..(.((((((((((((((..((((........))))....)))).))).)).))))).)..).....((((.......))))....)))...))..-- ( -28.20) >DroPse_CAF1 190428 105 - 1 ---CUACCUUUCUUCAGCAGAUUGGAGGCAUGCGGAUUGCGGCUCUUUGUGCUCAGCGAUGAGCAGUUCCAGCGAUGCCAUUGAAACUGUUGAGGAAGAGAGAGAU----------UG-- ---((..(((((((((((((.((.((((((((((((..((.((((.((((.....)))).)))).))))).)).))))).)).)).))))))))).))))..))..----------..-- ( -40.70) >DroWil_CAF1 226832 98 - 1 UACUUACCUUUCUUCAGCAGAUUGGAAGCAUGCGGAUUGCGACUUUUUGUGCUCAACGAUGAACAGUUCCAGCGAUGCCAUUGAAACUGGUGGG--------AAAA-------------- ..((((((((((....(((..((((((.(((.((....((.((.....))))....))))).....))))))...)))....))))..))))))--------....-------------- ( -22.60) >DroMoj_CAF1 225762 120 - 1 CACUUACCUUUCUUCAGCAGAUUGGAGGCGUGCGGAUUGCGGCUCUUUGUGCUCAGCGACGAGCAGUUCCAGCGAUGCCACUGAAACUGCAACCACAUAAAAUGAUUAGCUAAUUAUUGC ................((((.(..(.((((((((((..((.((((.((((.....)))).)))).))))).)).))))).)..)..))))..........(((((((....))))))).. ( -32.10) >DroPer_CAF1 192573 105 - 1 ---CUACCUUUCUUCAGCAGAUUGGAGGCAUGCGGAUUGCGGCUCUUUGUGCUCAGCGAUGAGCAGUUCCAGCGAUGCCAUUGAAACUGUUGAGGAAGAGAGAGAU----------UG-- ---((..(((((((((((((.((.((((((((((((..((.((((.((((.....)))).)))).))))).)).))))).)).)).))))))))).))))..))..----------..-- ( -40.70) >consensus _ACUUACCUUUCUUCAGCAGAUUGGAGGCAUGCGGAUUGCGGCUCUUUGUGCUCAGCGACGAGCAGUUCCAGCGAUGCCAUUGAAACUGUUGAG__A_A_A_AGAU__________UG__ .............(((((((.((.((((((((((((..((.((((.((((.....)))).)))).))))).)).))))).)).)).)))))))........................... (-21.93 = -23.72 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:26 2006