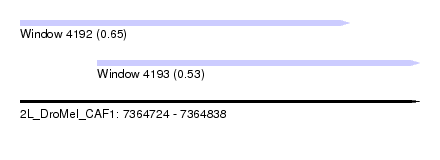
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,364,724 – 7,364,838 |
| Length | 114 |
| Max. P | 0.648302 |

| Location | 7,364,724 – 7,364,818 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.33 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
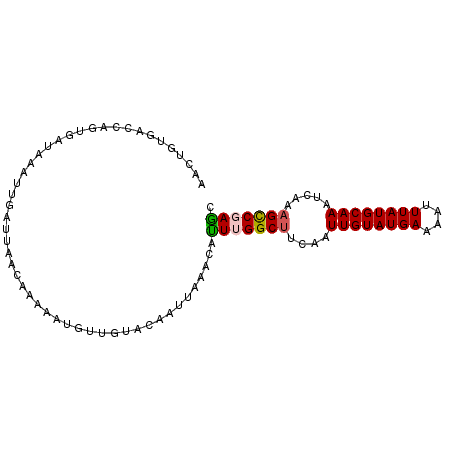
>2L_DroMel_CAF1 7364724 94 + 22407834 CACUGUGCCCAGUGAUAAAUUGAUUAACGAAAAUGCCGUGCAAUUAAACAUUUGGCCCCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAAC (((((....)))))...(((((....(((.......))).))))).....((((((.....((((((((....))))))))......)))))). ( -20.30) >DroVir_CAF1 204842 94 + 1 UUGUGUGUCCAGCAACAACAAAAGCAACAAAAAUGUGCUGCCACUAGAAUUUUGGCUUCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGC ....(((..(((((.((................))))))).)))......((((((((...((((((((....))))))))....)))))))). ( -20.79) >DroPse_CAF1 184760 94 + 1 AAUUGAGAGCAGUGAUAAAUUGAUUAACAAAAAUGUUGUACAAUUAAACAUUUGGCUACAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGC ........((.((....(((((..(((((....)))))..)))))..))..((((((....((((((((....)))))))).....)))))))) ( -17.90) >DroGri_CAF1 173113 94 + 1 UUAUGUGGACAGCAACAACAAAAGAAACAAAAAUGUUCUUCAACUAAAAUUUAGGCUUCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGUCGAGC .......((((((........(((((.((....)))))))...(((.....))))))....((((((((....))))))))......))).... ( -12.60) >DroAna_CAF1 158997 94 + 1 AACAGUGCCAAGUGAUAAAUUGAUUAACGAAAAUGCUUUACAAUUAAACACUGGGCUUCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGC ..(((((....((((...(((..........)))...)))).......)))))(((((...((((((((....))))))))....))))).... ( -17.90) >DroPer_CAF1 186868 94 + 1 AAUUGAGAGCAGUGAUAAAUUGAUUAACAAAAAUGUUGUACAAUUAAACAUUUGGCUACAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGC ........((.((....(((((..(((((....)))))..)))))..))..((((((....((((((((....)))))))).....)))))))) ( -17.90) >consensus AACUGUGACCAGUGAUAAAUUGAUUAACAAAAAUGUUGUACAAUUAAACAUUUGGCUUCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGC ..................................................(((((((....((((((((....)))))))).....))))))). (-11.25 = -11.33 + 0.09)

| Location | 7,364,746 – 7,364,838 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
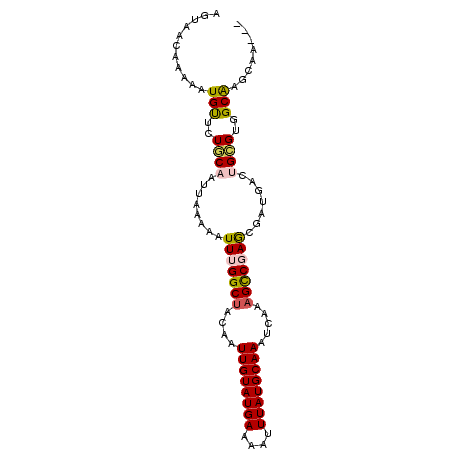
>2L_DroMel_CAF1 7364746 92 + 22407834 AUUAACGAAAAUGCCGUGCAAUUAAACAUUUGGCCCCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAACGAUGACUGCGUGGCGAGCAA--- ...........(((((((((.(((....((((((.....((((((((....))))))))......))))))...))).))))))))).....--- ( -23.30) >DroVir_CAF1 204864 92 + 1 AGCAACAAAAAUGUGCUGCCACUAGAAUUUUGGCUUCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGCGAUGACUGCGUGGCAAGCAA--- .............(((((((((.......(((((((...((((((((....))))))))....)))))))(((.....)))))))).)))).--- ( -29.30) >DroPse_CAF1 184782 95 + 1 AUUAACAAAAAUGUUGUACAAUUAAACAUUUGGCUACAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGCGAUGACUGUGUGGCAAGAAGCAA ...........(((..((((.(((....(((((((....((((((((....)))))))).....)))))))...))).))))..)))........ ( -21.90) >DroGri_CAF1 173135 92 + 1 AGAAACAAAAAUGUUCUUCAACUAAAAUUUAGGCUUCAAUUGUAUGAAAAUUUAUGCAAAUCAAAGUCGAGCGAUGACUGCGUGGCAAGCAA--- ...........((((................(((((...((((((((....))))))))....)))))..((.(((....))).)).)))).--- ( -15.30) >DroMoj_CAF1 217538 92 + 1 UGCAAUCAAAAUGUGCUGCUAGUCAAAUUUGGGCUACAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGCGAUGACUGCGUGGCGAGCAA--- (((..........(((..(((((((...((.((((....((((((((....)))))))).....)))).))...)))))).)..))).))).--- ( -25.20) >DroPer_CAF1 186890 95 + 1 AUUAACAAAAAUGUUGUACAAUUAAACAUUUGGCUACAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGCGAUGACUGUGUGGCAAGAAGCAA ...........(((..((((.(((....(((((((....((((((((....)))))))).....)))))))...))).))))..)))........ ( -21.90) >consensus AGUAACAAAAAUGUUCUGCAAUUAAAAAUUUGGCUACAAUUGUAUGAAAAUUUAUGCAAAUCAAAGCCGAGCGAUGACUGCGUGGCAAGCAA___ ...........(((..((((........(((((((....((((((((....)))))))).....))))))).......))))..)))........ (-15.35 = -15.38 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:24 2006