
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,339,679 – 7,339,775 |
| Length | 96 |
| Max. P | 0.762391 |

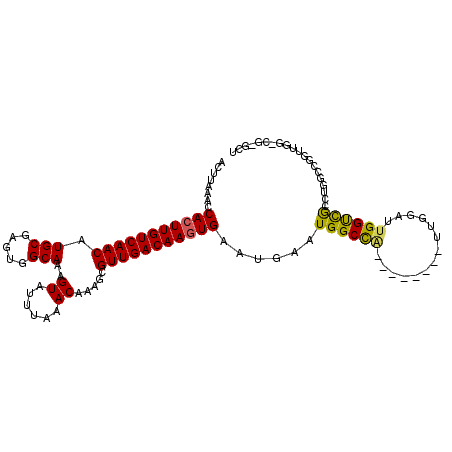
| Location | 7,339,679 – 7,339,775 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -23.12 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 7339679 96 - 22407834 ACUUAAACCACUUGUCAACAUGCGAGUGGCAAAGUAUUUAAACAAAGCGUUGACAAGUGAAUGAAUGGCCA--------UUGGAUUGGUUGCCUGGCCGG------G-GCU ........(((((((((((.(((.....)))..((......)).....)))))))))))......((((((--------..((........)))))))).------.-... ( -29.10) >DroVir_CAF1 181490 110 - 1 ACUUAAACCACUUGUCAACAUGCGAGUGGCAAAGUAUUUAAACAAAGCGUUGACAACUGAAUGAAUGGCUGGCCGCUCGUUCAGUUGGCCAACAGAUCGUUUGGGCC-GCU ((((...((((((((......))))))))..))))..........((((....(((((((((((.(((....))).)))))))))))(((.((.....))...))))-))) ( -38.70) >DroGri_CAF1 152096 102 - 1 ACUUAAACCACUUGUCAACAUGCGAGUGGCAAAGUAUUUAAACAAAGCGUUGACAAGUGAAUGAAUGGCC--------GCUCAGUUGGCCAACUGAUCGUUUGGGCC-GCG .((((((((((((((((((.(((.....)))..((......)).....)))))))))))......(((((--------(......)))))).......)))))))..-... ( -33.20) >DroEre_CAF1 135178 102 - 1 ACUUAAACCACUUGUCAACAUGCGAGUGGCAAAGUAUUUAAACAAAGCGUUGACAAGUGAAUGAAUGGCCA--------UUGGAUUGGUCGGCUGGUCGGCUGGCCG-GCU ........(((((((((((.(((.....)))..((......)).....))))))))))).......(((((--------......)))))((((((((....)))))-))) ( -37.70) >DroYak_CAF1 137617 103 - 1 ACUUAAACCACUUGUCAACAUGCGAGUGGCAAAGUAUUUAAACAAAGCGUUGACAAGUGAAUGAAUGGCCA--------UUGGAUUGGUCGCCUGGCCGGUCGGCGGUGCU ........(((((((((((.(((.....)))..((......)).....))))))))))).......(((((--------......)))))((.(.(((....))).).)). ( -34.00) >DroAna_CAF1 140156 94 - 1 ACUUAAACCACUUGUCAACAUGCGAGUUGCAAAGUAUUUAAACAAAGCGUUGACAAGUGAAUGAAUGGCCA--------UUGGAUUGGUCGCCAGGCUG---------CGA ........(((((((((((.(((.....)))..((......)).....))))))))))).......(((((--------......)))))((......)---------).. ( -26.40) >consensus ACUUAAACCACUUGUCAACAUGCGAGUGGCAAAGUAUUUAAACAAAGCGUUGACAAGUGAAUGAAUGGCCA________UUGGAUUGGUCGCCUGGCCGGUUGG_CG_GCU ........(((((((((((.(((.....)))..((......)).....)))))))))))......((((((..............)))))).................... (-23.12 = -22.62 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:18 2006